Electronic Supplementary Information S9
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

METACYC ID Description A0AR23 GO:0004842 (Ubiquitin-Protein Ligase
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Heat Stress Responsive Zostera marina Genes, Southern Population (α=0. -

Peroxidase Activity As an Indicator of the Iron Deficiency in Banana
IndianJ Plant Physiol., Vol. 5, No.4, (N.S.) pp. 389-391 (Oct.-Dec., 2000) SHORT COMMUNICATION PEROXIDASE ACTIVITY AS AN INDICATOR OF THE IRON DEFICIENCY IN BANANA K. BALAKRISHNAN Department ofCrop Physiology, Horticultural College and Research Institute, Periyakulam - 625 604 Received on 30 Sept., 1998, Revised on 30 Nov., 2000 Effect oflime induced iron chlorosis on enzyme activities was studied in Banana cv. Rasthali. Iron content decreased progressively as the intensity of chlorosis increased. Iron content had positive correlation with catalase, peroxidase, acid phosphatase, polyphenol oxidase and nitrate reductase. Among the enzymes, the activityofperoxidasehad highest positivesignificant(r=997**) associationwith Fecontent. Hence, peroxidase activity could serve as a diagnostic tool to indentify the Fe deficiency/Fe status in Banana. Key words: Banana, chlorosis, iron, peroxidase Among the micrountrients, Fe deficiency is the most ofgreenness in sixmonths old crop. This leafwas used for widespread in ourcountry(Takkar, 1996). Iron deficiency all the physiological analysis viz chlorophyll (Yoshida et is very common in calcareous soils and hence termed as al., 1976) chlorophyllase (Almela et al., 1990), catalase, lime induced iron chlorosis. In a normal green plant 60% peroxidase and polyphenol oxidase (Kar and Mishra, ofall leafiron is present in the chlorophyIl and hence any 1976), acid phosphatase (Parida and Mishra, 1980) and reduction in Fe contentcauses chlorosis (Chen and Barak, nitrate reductase (Klepperetal., 1973). The Fe content of 1982). Iron deficiency in plant not only causes chlorosis the plant sample was estimated using atomic absorption because of its involvement in chlorophyll synthesis, but spectrophotometry. Data were subjected to simple also reduces the activity ofcertain enzymes viz., catalase correlation co-efficient. -

Allele-Specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish
Allele-specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish by Ailu Chen A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama August 1, 2015 Keywords: catfish, interspecific hybrids, allele-specific expression, ribosomal protein Copyright 2015 by Ailu Chen Approved by Zhanjiang Liu, Chair, Professor, School of Fisheries, Aquaculture and Aquatic Sciences Nannan Liu, Professor, Entomology and Plant Pathology Eric Peatman, Associate Professor, School of Fisheries, Aquaculture and Aquatic Sciences Aaron M. Rashotte, Associate Professor, Biological Sciences Abstract Interspecific hybridization results in a vast reservoir of allelic variations, which may potentially contribute to phenotypical enhancement in the hybrids. Whether the allelic variations are related to the downstream phenotypic differences of interspecific hybrid is still an open question. The recently developed genome-wide allele-specific approaches that harness high- throughput sequencing technology allow direct quantification of allelic variations and gene expression patterns. In this work, I investigated allele-specific expression (ASE) pattern using RNA-Seq datasets generated from interspecific catfish hybrids. The objective of the study is to determine the ASE genes and pathways in which they are involved. Specifically, my study investigated ASE-SNPs, ASE-genes, parent-of-origins of ASE allele and how ASE would possibly contribute to heterosis. My data showed that ASE was operating in the interspecific catfish system. Of the 66,251 and 177,841 SNPs identified from the datasets of the liver and gill, 5,420 (8.2%) and 13,390 (7.5%) SNPs were identified as significant ASE-SNPs, respectively. -

Phospholipases, Nucleic Acids Encoding Them and Methods for Making and Using Them
(19) TZZ¥_Z_ _T (11) EP 3 190 182 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 12.07.2017 Bulletin 2017/28 C12N 9/20 (2006.01) C12N 1/20 (2006.01) C12N 15/00 (2006.01) C07H 21/04 (2006.01) (21) Application number: 16184319.8 (22) Date of filing: 08.03.2005 (84) Designated Contracting States: • FIELDING, Roderick AT BE BG CH CY CZ DE DK EE ES FI FR GB GR San Diego, CA 92109 (US) HU IE IS IT LI LT LU MC NL PL PT RO SE SI SK TR • BROWN, Robert C. San Diego, CA 92130 (US) (30) Priority: 08.03.2004 US 796907 • VASAVADA, Amit Poway, CA 92064 (US) (62) Document number(s) of the earlier application(s) in • TAN, Xuqiu accordance with Art. 76 EPC: San Diego, CA 92130 (US) 05727242.9 / 1 748 954 • BADILLO, Adrian Poway, CA 92064 (US) (27) Previously filed application: • VAN HOEK, Wilhelmus P. 08.03.2005 PCT/US2005/007908 San Diego, CA 92126 (US) • JANSSEN, Giselle (71) Applicant: DSM IP Assets B.V. San Diego, CA 92121 (US) 6411 TE Heerlen (NL) • ISAAC, Charles Carlsbad, CA 92008 (US) (72) Inventors: • BURK, Mark J. • GRAMATIKOVA, Svetlana San Diego, CA 92130 (US) San Diego, CA 92122 (US) • HAZLEWOOD, Geoff (74) Representative: Cazemier, Anne Engeline et al San Diego, CA 92130 (US) DSM Intellectual Property •LAM,David P.O. Box 4 Encinitas, CA 92024 (US) 6100 AA Echt (NL) • BARTON, Nelson R. San Diego, CA 92131 (US) Remarks: • STURGIS, Blake G. •A request for correction of the description has been Solana Beach, CA 92075 (US) filed pursuant to Rule 139 EPC. -

Fruit Ripening and Storage
OPEN Citation: Horticulture Research (2014) 1, 6; doi:10.1038/hortres.2014.6 ß 2014 Nanjing Agricultural University All rights reserved 2052-7276/14 www.nature.com/hortres ARTICLE Dynamic changes in proteins during apple (Malus x domestica) fruit ripening and storage Yun Shi1, Li Jiang1, Li Zhang2, Ruoyi Kang1 and Zhifang Yu1 A proteomic study, using two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight/time-of-flight, was conducted in apple fruit (cv. ‘Golden Delicious’) starting at 10 days prior to harvest through 50 days in storage. Total protein was extracted using a phenol/sodium dodecyl sulfate protocol. More than 400 protein spots were detected in each gel and 55 differentially expressed proteins (p,0.05) were subjected to matrix-assisted laser desorption/ionization time-of-flight/ time-of-flight analysis. Fifty-three of these proteins were finally identified using an apple expressed sequence tag database downloaded from Genome Database for Rosaceae and placed into six categories. The categories and the percentage of proteins placed in each category were stress response and defense (49.0%), energy and metabolism (34.0%), fruit ripening and senescence (5.6%), signal transduction (3.8%), cell structure (3.8%) and protein synthesis (3.8%). Proteins involved in several multiple metabolic pathways, including glycolysis, pentose–phosphate pathway, anti-oxidative systems, photosynthesis and cell wall synthesis, were downregulated, especially during the climacteric burst in respiration and during the senescent stages of fruit development. Proteins classified as allergens or involved in cell wall degradation were upregulated during the ripening process. Some protein spots exhibited a mixed pattern (increasing to maximal abundance followed by a decrease), such as 1-aminocyclopropane-1-carboxylate oxidase, L-ascorbate peroxidase and abscisic acid response proteins. -

Genome-Wide Analysis of Glyoxalase-Like Gene Families in Grape
Li et al. BMC Genomics (2019) 20:362 https://doi.org/10.1186/s12864-019-5733-y RESEARCHARTICLE Open Access Genome-wide analysis of glyoxalase-like gene families in grape (Vitis vinifera L.) and their expression profiling in response to downy mildew infection Tiemei Li1,2,3, Xin Cheng1,2,3, Yuting Wang1,2,3, Xiao Yin1,2,3, Zhiqian Li1,2,3, Ruiqi Liu1,2,3, Guotian Liu1,2,3, Yuejin Wang1,2,3 and Yan Xu1,2,3* Abstract Background: The glyoxalase system usually comprises two enzymes, glyoxalase I (GLYI) and glyoxalase II (GLYII). This system converts cytotoxic methylglyoxal (MG) into non-toxic D-lactate in the presence of reduced glutathione (GSH) in two enzymatic steps. Recently, a novel type of glyoxalase III (GLYIII) activity has observed in Escherichia coli that can detoxify MG into D-lactate directly, in one step, without a cofactor. Investigation of the glyoxalase enzymes of a number of plant species shows the importance of their roles in response both to abiotic and to biotic stresses. Until now, glyoxalase gene families have been identified in the genomes of four plants, Arabidopsis, Oryza sativa, Glycine max and Medicago truncatula but no similar study has been done with the grapevine Vitis vinifera L. Results: In this study, four GLYI-like,twoGLYII-like and three GLYIII-like genesareidentifiedfromthegenomedatabaseof grape. All these genes were analysed in detail, including their chromosomal locations, phylogenetic relationships, exon-intron distributions, protein domain organisations and the presence of conserved binding sites. Using quantitative real-time PCR analysis (qRT-PCR), the expression profiles of these geneswereanalysedindifferent tissues of grape, and also when under infection stress from downy mildew (Plasmopara viticola). -

Legionella Genus Genome Provide Multiple, Independent Combinations for Replication in Human Cells
Supplemental Material More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells Laura Gomez-Valero1,2, Christophe Rusniok1,2, Danielle Carson3, Sonia Mondino1,2, Ana Elena Pérez-Cobas1,2, Monica Rolando1,2, Shivani Pasricha4, Sandra Reuter5+, Jasmin Demirtas1,2, Johannes Crumbach1,2, Stephane Descorps-Declere6, Elizabeth L. Hartland4,7,8, Sophie Jarraud9, Gordon Dougan5, Gunnar N. Schroeder3,10, Gad Frankel3, and Carmen Buchrieser1,2,* Table S1: Legionella strains analyzed in the present study Table S2: Type IV secretion systems predicted in the genomes analyzed Table S3: Eukaryotic like domains identified in the Legionella proteins analyzed Table S4: Small GTPases domains detected in the genus Legionella as defined in the CDD ncbi domain database Table S5: Eukaryotic like proteins detected in the Legionella genomes analyzed in this study Table S6: Aminoacid identity of the Dot/Icm components in Legionella species with respect to orthologous proteins in L. pneumophila Paris Table S7: Distribution of seventeen highly conserved Dot/Icm secreted substrates Table S8: Comparison of the effector reperotoire among strains of the same Legionella species Table S9. Number of Dot/Icm secreted proteins predicted in each strain analyzed Table S10: Replication capacity of the different Legionella species analyzed in this study and collection of literature data on Legionella replication Table S11: Orthologous table for all genes of the 80 analyzed strains based on PanOCT. The orthologoss where defined with the program PanOCT using the parameters previously indicated in material and methods.) Figure S1: Distribution of the genes predicted to encode for the biosynthesis of flagella among all Legionella species. -

1 Polymorphisms in Brucella Carbonic Anhydrase II Mediate CO2 Dependence and Fitness 1 in Vivo. 2 3 García-Lobo JM1, Ortiz Y1
bioRxiv preprint doi: https://doi.org/10.1101/804740; this version posted October 15, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Polymorphisms in Brucella Carbonic anhydrase II mediate CO2 dependence and fitness 2 in vivo. 3 4 García-Lobo JM1, Ortiz Y1, González-Riancho C1, Seoane A1, Arellano-Reynoso B2, and 5 Sangari FJ1* 6 7 *Corresponding author 8 1. Instituto de Biomedicina y Biotecnología de Cantabria (IBBTEC), CSIC-Universidad de 9 Cantabria, and Departamento de Biología Molecular, Universidad de Cantabria, 39011 10 Santander, Spain. 11 2. Departamento de Microbiología, Facultad de Medicina Veterinaria y Zootecnia, Universidad 12 Nacional Autónoma de México, Circuito Exterior de Ciudad Universitaria, Delegación 13 Coyoacán, Mexico City, C.P. 04510, Mexico. 14 15 FJS conceived and coordinated the study, conducted bacteriology work and wrote the 16 manuscript. JMGL analyzed the data and wrote the manuscript. YO, CGR, AS and BAR 17 conducted bacteriology work. All authors interpreted the data, corrected the manuscript, and 18 approved the content for publication. 19 20 21 Keywords. Brucella, Carbonic anhydrase, CO2 requirement, fitness, protein structure 22 1 bioRxiv preprint doi: https://doi.org/10.1101/804740; this version posted October 15, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 23 Abstract 24 25 Some Brucella isolates are known to require an increased concentration of CO2 for growth, 26 especially in the case of primary cultures obtained directly from infected animals. -

Supplementary Figure S1 Functional Characterisation of Snmp:GFP
doi: 10.1038/nature06328 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE LEGENDS Figure S1 | Functional characterisation of SNMP fusion proteins. Dose-response curve for cVA in Or67d neurons of wild-type (Berlin), SNMP mutant (Or67d- GAL4/+;SNMP1/SNMP2), SNMP:GFP rescue (Or67d-GAL4/UAS- SNMP:GFP;SNMP1/SNMP2) and YFP(1):Or83b/SNMP:YFP(2) rescue (Or67d:GAL4,UAS-YFP(1):Or83b/UAS-SNMP:YFP(2);SNMP1,Or83b2/SNMP2,Or83b1 ) animals. Mean responses are plotted (± s.e.m; wild-type n=47, SNMP mutant n=46, SNMP:GFP rescue n=20; YFP(1):Or83b/SNMP:YFP(2) rescue n=22; ≤4 sensilla/animal, mixed genders). Wild-type and SNMP:GFP rescue responses to cVA are not significantly different (ANOVA; p>0.1175). YFP(1):Or83b/SNMP:YFP(2) rescue responses to cVA are highly significantly different from SNMP mutants and from wild- type (ANOVA; p<0.0001), indicating partial rescue. Figure S2 | Cell type-specific rescue of SNMP expression. a, Immunostaining for mCD8:GFP (anti-GFP, green) and LUSH (magenta) in LUSH-GAL4/UAS-mCD8:GFP animals reveals faithful recapitulation of endogenous expression by the LUSH-GAL4 driver. b, Two-colour RNA in situ hybridisation for SNMP (green) and Or67d (magenta) in antennal sections of wild-type, Or67d neuron SNMP rescue (Or67d-GAL4/UAS- SNMP;SNMP1/SNMP2) and support cell SNMP rescue (LUSH-GAL4/UAS- SNMP;SNMP1/SNMP2) animals. www.nature.com/nature 1 Benton et al., Figure S1 ) -1 wild-type 120 SNMP:GFP rescue 80 YFP(1):Or83b/SNMP:YFP(2) rescue 40 Corrected response (spikes s 0 SNMP-/- 0 0.1 1 10 100 cVA (%) www.nature.com/nature 2 Benton -

Aberrant Expression of Tetraspanin Molecules in B-Cell Chronic Lymphoproliferative Disorders and Its Correlation with Normal B-Cell Maturation
Leukemia (2005) 19, 1376–1383 & 2005 Nature Publishing Group All rights reserved 0887-6924/05 $30.00 www.nature.com/leu Aberrant expression of tetraspanin molecules in B-cell chronic lymphoproliferative disorders and its correlation with normal B-cell maturation S Barrena1,2, J Almeida1,2, M Yunta1,ALo´pez1,2, N Ferna´ndez-Mosteirı´n3, M Giralt3, M Romero4, L Perdiguer5, M Delgado1, A Orfao1,2 and PA Lazo1 1Instituto de Biologı´a Molecular y Celular del Ca´ncer, Centro de Investigacio´n del Ca´ncer, Consejo Superior de Investigaciones Cientı´ficas-Universidad de Salamanca, Salamanca, Spain; 2Servicio de Citometrı´a, Universidad de Salamanca and Hospital Universitario de Salamanca, Salamanca, Spain; 3Servicio de Hematologı´a, Hospital Universitario Miguel Servet, Zaragoza, Spain; 4Hematologı´a-hemoterapia, Hospital Universitario Rı´o Hortega, Valladolid, Spain; and 5Servicio de Hematologı´a, Hospital de Alcan˜iz, Teruel, Spain Tetraspanin proteins form signaling complexes between them On the cell surface, tetraspanin antigens are present either as and with other membrane proteins and modulate cell adhesion free molecules or through interaction with other proteins.25,26 and migration properties. The surface expression of several tetraspanin antigens (CD9, CD37, CD53, CD63, and CD81), and These interacting proteins include other tetraspanins, integri- F 22,27–30F their interacting proteins (CD19, CD21, and HLA-DR) were ns particularly those with the b1 subunit HLA class II 31–33 34,35 analyzed during normal B-cell maturation and compared to a moleculesFeg HLA DR -, CD19, the T-cell recep- group of 67 B-cell neoplasias. Three patterns of tetraspanin tor36,37 and several other members of the immunoglobulin expression were identified in normal B cells. -
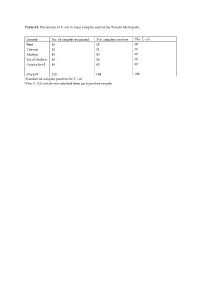
Supplementary File 1
Table S1. Prevalence of E. coli in meat samples sold at the Tamale Metropolis. Sample No. of samples examined aNo. samples positive bNo. E. coli Beef 45 39 39 Chevon 45 34 34 Mutton 45 40 40 Local chicken 45 36 36 Guinea fowl 45 40 40 Overall 225 189 189 aNumber of samples positive for E. coli. bOne E. Coli isolate was selected from each positive sample. Table S2. A table showing the eBURST (Based Upon Related Sequence Types) analyses of the study sequence types with global curated STs in Escherichia PubMLST database. MLST (Isolate) Type of clone Closet global ancestry Source sequence type (ST) ST69 (SG6) Similar a ST69 Animal (Food), Human ST155 (SLC2, Similar ST155 Animal (Food), Human, TLC13, CM4) Environment ST297 (TLC1) Similar ST297 Human ST1727 (NC3) Similar ST1727 Human ST44 (AC1) Single-Locus Variant ST10, ST752 Animal (Food), (SLV) b Human ST469 (CC6) Single-Locus Variant ST162 Food (SLV) ST540 (AB1, Single-Locus Variant ST4093 Human TG1) (SLV) ST1141 (NM11) Single-Locus Variant ST10, ST744 Animal (Food), (SLV) Human ST7473 (NB12) Single-Locus Variant ST10 Animal (Food), (SLV) Human ST6646 (CB1) Satellite c None - ST7483 (NB12) Satellite None - a Similar: study isolate was similar to a global curated known sequence type. b Single-Locus Variant (SLV): study isolate only shared similarity with global curated known sequence types that differed in one allelic gene. c Satellite: study isolate as a distantly related and did not shared any similarity with global curated known sequence types. Table S3. In silico identification and characterization of conserved stress response mechanisms in the E. -

The Tetraspanin TSPAN33 Controls TLR-Triggered Macrophage Activation Through Modulation of NOTCH Signaling
The Tetraspanin TSPAN33 Controls TLR-Triggered Macrophage Activation through Modulation of NOTCH Signaling This information is current as Almudena Ruiz-García, Susana López-López, José Javier of September 25, 2021. García-Ramírez, Victoriano Baladrón, María José Ruiz-Hidalgo, Laura López-Sanz, Ángela Ballesteros, Jorge Laborda, Eva María Monsalve and María José M. Díaz-Guerra J Immunol published online 29 August 2016 Downloaded from http://www.jimmunol.org/content/early/2016/08/27/jimmun ol.1600421 Supplementary http://www.jimmunol.org/content/suppl/2016/08/27/jimmunol.160042 http://www.jimmunol.org/ Material 1.DCSupplemental Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 25, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published August 29, 2016, doi:10.4049/jimmunol.1600421 The Journal of Immunology The Tetraspanin TSPAN33 Controls TLR-Triggered Macrophage Activation through Modulation of NOTCH Signaling Almudena Ruiz-Garcı´a,1 Susana Lo´pez-Lo´pez,1 Jose´ Javier Garcı´a-Ramı´rez, Victoriano Baladro´n, Marı´a Jose´ Ruiz-Hidalgo, Laura Lo´pez-Sanz, A´ ngela Ballesteros, Jorge Laborda, Eva Marı´a Monsalve, and Marı´a Jose´ M.