Navigating Metabolism
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Effect of an Oral Or Intravenous Nucleotide-Free Diet on Selected Enzyme Activities in Purine Metabolism in Rats
RICE UNIVERSITY THE EFFECT OF AN ORAL OR INTRAVENOUS NUCLEOTIDE-FREE DIET ON SELECTED ENZYME ACTIVITIES IN PURINE METABOLISM IN RATS. by SANDRA LYNN SNYDER A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE MASTER OF ARTS APPROVED, THESIS COMMITTEE: .T Dr. Frederi Associate Professor of B Chemistry Dr. Susan M. B^fget Assistant Professor of Biochemistry Dr. George J. Æchfoépfer Professor of Biochemistry and Chemistry Dr. George'N. Bennett Assistant Professor of Biochemistry Dr. Roger L. Storck Professor of Biology HOUSTON, TEXAS MARCH, 1982 ABSTRACT THE EFFECT OF AN ORAL OR INTRAVENOUS NUCLEOTIDE-FREE DIET ON SELECTED ENZYME ACTIVITIES IN PURINE METABOLISM IN RATS SANDRA LYNN SNYDER Interrelationships between purine metabolism and immunity, cancer, and the diet have been considered. In studying trends of metabolic changes which occur in response to changes in the purine content of the diet, it has been hypothesized that when purines are lacking from the diet, there is a general shift from a catabolic to an anabolic state. In the present investigation, the activities of selected enzymes on purine metabolism in rats were studied with respect to an oral or intravenous nucleotide-free diet compared to the activities in rats fed normal chow. The intravenous nucleotide-free diet caused a decrease in purine nucleoside phosphorylase activity, an increase in adenine phosphoribosyl transferase, and no change in the activity of hypoxanthine-guanine phosphoribosyl transferase, with respect to a normal control diet. The orally fed nucleotide-free diet caused a decrease in the activity of purine nucleoside phosphorylase and xanthine oxidase and increase in pancreatic ribonucléase and no change in adenine phosphoribosyl transferase or hyphoxanthine-guanine phosphoribosyl transferase, with respect to a normal control diet. -

Is Human Hibernation Possible?
ANRV334-ME59-12 ARI 16 December 2007 14:50 Is Human Hibernation Possible? Cheng Chi Lee Department of Biochemistry and Molecular Biology, University of Texas Health Science Center, Houston, Texas 77030; email: [email protected] Annu. Rev. Med. 2008. 59:177–86 Key Words The Annual Review of Medicine is online at hypothermia, 5-AMP, torpor, hypometabolism http://med.annualreviews.org This article’s doi: Abstract 10.1146/annurev.med.59.061506.110403 The induction of hypometabolism in cells and organs to reduce is- Copyright c 2008 by Annual Reviews. chemia damage holds enormous clinical promise in diverse fields, in- All rights reserved cluding treatment of stroke and heart attack. However, the thought 0066-4219/08/0218-0177$20.00 that humans can undergo a severe hypometabolic state analogous to hibernation borders on science fiction. Some mammals can enter a severe hypothermic state during hibernation in which metabolic activity is extremely low, and yet full viability is restored when the animal arouses from such a state. To date, the underlying mecha- nism for hibernation or similar behaviors remains an enigma. The beneficial effect of hypothermia, which reduces cellular metabolic demands, has many well-established clinical applications. However, severe hypothermia induced by clinical drugs is extremely difficult and is associated with dramatically increased rates of cardiac arrest for nonhibernators. The recent discovery of a biomolecule, 5-AMP, which allows nonhibernating mammals to rapidly and safely enter severe hypothermia could remove this impediment and enable the wide adoption of hypothermia as a routine clinical tool. 177 ANRV334-ME59-12 ARI 16 December 2007 14:50 INTRODUCTION ing mammals. -

Enhancing Nucleotide Metabolism Protects Against Mitochondrial Dysfunction and Neurodegeneration in a PINK1 Model of Parkinson’S Disease
ARTICLES Enhancing nucleotide metabolism protects against mitochondrial dysfunction and neurodegeneration in a PINK1 model of Parkinson's disease Roberta Tufi1, Sonia Gandhi2, Inês P. de Castro1, Susann Lehmann1, Plamena R. Angelova2, David Dinsdale1, Emma Deas2, Hélène Plun-Favreau2, Pierluigi Nicotera3, Andrey Y. Abramov2, Anne E. Willis1, Giovanna R. Mallucci1, Samantha H. Y. Loh1,4 and L. Miguel Martins1,4 Mutations in PINK1 cause early-onset Parkinson's disease (PD). Studies in Drosophila melanogaster have highlighted mitochondrial dysfunction on loss of Pink1 as a central mechanism of PD pathogenesis. Here we show that global analysis of transcriptional changes in Drosophila pink1 mutants reveals an upregulation of genes involved in nucleotide metabolism, critical for neuronal mitochondrial DNA synthesis. These key transcriptional changes were also detected in brains of PD patients harbouring PINK1 mutations. We demonstrate that genetic enhancement of the nucleotide salvage pathway in neurons of pink1 mutant flies rescues mitochondrial impairment. In addition, pharmacological approaches enhancing nucleotide pools reduce mitochondrial dysfunction caused by Pink1 deficiency. We conclude that loss of Pink1 evokes the activation of a previously unidentified metabolic reprogramming pathway to increase nucleotide pools and promote mitochondrial biogenesis. We propose that targeting strategies enhancing nucleotide synthesis pathways may reverse mitochondrial dysfunction and rescue neurodegeneration in PD and, potentially, other diseases linked to mitochondrial impairment. The role of mitochondrial impairment in PD has long been debated. By combining transcriptional and metabolic profiling, we have Recently, the identification of causative mutations in PINK1, a gene uncovered significant alterations in the nucleotide metabolism encoding a mitochondrial kinase in PD patients has renewed interest in networks of pink1 mutant flies. -

1 Targeting Subtype-Specific Metabolic Preferences in Nucleotide
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.19.049577; this version posted May 9, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Targeting subtype-specific metabolic preferences in nucleotide biosynthesis inhibits tumor growth in a breast cancer model Martin P. Ogrodzinski1,2, Shao Thing Teoh1, and Sophia Y. Lunt1,3* 1Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI, USA 2Department of Physiology, Michigan State University, East Lansing, MI, USA 3Department of Chemical Engineering and Materials Science, Michigan State University, East Lansing, MI, USA *Correspondence: Sophia Y. Lunt, Ph.D. Biochemistry Building 603 Wilson Rd Room 522A Michigan State University East Lansing, MI 48824, USA Phone: 517-432-4886 Email: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.04.19.049577; this version posted May 9, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. ABSTRACT Investigating metabolic rewiring in cancer can lead to the discovery of new treatment strategies for breast cancer subtypes that currently lack targeted therapies. Using MMTV- Myc driven tumors to model breast cancer heterogeneity, we investigated metabolic differences between two histological subtypes, the epithelial-mesenchymal transition (EMT) and the papillary subtypes, using a combination of genomic and metabolomic techniques. We identified differences in nucleotide metabolism between EMT and papillary subtypes: EMT tumors preferentially use the nucleotide salvage pathway, while papillary tumors prefer de novo nucleotide biosynthesis. -
When the Reaction Is
Table S3. iJL1678-ME model modification (blocked reactions) Iter. Cat. ID Name Formula Subsystem Comments (When the reaction is turned on) 1 bp2 EDD 6-phosphogluconate dehydratase 6pgc_c⇌2ddg6p_c + h2o_c Pentose Phosphate Pathway Create a major effect of steep acetate overflow elevation in high growth. Comparing to the main glycolytic pathway, it is metabolicly less efficient but proteomicly more efficient. bp1 ICL Isocitrate lyase icit_c→glx_c + succ_c Anaplerotic Reactions Bypass for the main TCA cycle pathways from turning isocitrate to succinate, when ICL is turned on, Isocitrate dehydrogenase(ICDHyr), 2-Oxogluterate dehydrogenase(AKGDH) and Succinyl-CoA synthetase (ATP-forming,SUCOAS) would reduce. Ref. (1) and (2) shows that this reaction is off in higher growth. Ref. (3) shows that this reaction is converging to being off when the dynamic of respiration using enzyme kinetics is simulated. 2 bp1 ABTA 4-aminobutyrate transaminase 4abut_c + akg_c⇌glu__L_c + sucsal_c Arginine and Proline Metabolism Another backup pathway of succinate production, from 2-Oxoglutarate (akg). Respiration would be induced when it is on, since the flux through ETC(CYTBO3_4pp and ATPS4rpp) would increase. As it requires the co-factor pyridoxal 5'-phosphate(2−) to get catalyzed(4), indicating that this reaction is regulated by the flux of other reactions(pyridoxal 5'- phosphate(2-) production, etc.). 3 GLYAT Glycine C-acetyltransferase accoa_c + gly_c⇌2aobut_c + coa_c Glycine and Serine Metabolism A reaction that back up for the respiration. Reactions fluxes in TCA cycle would drop when this reaction is turned on. It also requires pyridoxal 5'-phosphate(2−) for the regulation. 4 NADTRHD NAD transhydrogenase nad_c + nadph_c⇌nadh_c + nadp_c Oxidative Phosphorylation A reaction that would make the transition between NAD and NADP metabolically more efficient. -

Emerging Roles of Nucleotide Metabolism in Cancer Development: Progress and Prospect
www.aging-us.com AGING 2021, Vol. 13, No. 9 Review Emerging roles of nucleotide metabolism in cancer development: progress and prospect Jingsong Ma1,2, Mengya Zhong1,2, Yubo Xiong1,2, Zhi Gao3, Zhengxin Wu4, Yu Liu5, Xuehui Hong1,2 1Institute of Gastrointestinal Oncology, School of Medicine, Xiamen University, Fujian, Xiamen 361000, China 2Department of Gastrointestinal Surgery, Zhongshan Hospital, Xiamen University, Fujian, Xiamen 361000, China 3National Center for International Research of Biological Targeting Diagnosis and Therapy, Guangxi Key Laboratory of Biological Targeting Diagnosis and Therapy Research, Guangxi Medical University, Guangxi, Nanning 53000, China 4Medical College of Guangxi University, Guangxi, Nanning 530000, China 5General Surgery Center, Bazhong Central Hospital, Sichuan, Bazhong 636000, China Correspondence to: Xuehui Hong; email: [email protected] Keywords: nucleotide metabolism, tumor immunity, key metabolic enzyme, signaling pathway, oncogene-induced senescence Received: August 12, 2020 Accepted: March 29, 2021 Published: May 5, 2021 Copyright: © 2021 Ma et al. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Abnormal cancer metabolism occurs throughout the development of tumors. Recent studies have shown that abnormal nucleotide metabolism not only accelerates the development of tumors but also inhibits the normal immune response in the tumor microenvironment. Although few relevant experiments and reports are available, study of the interaction between nucleotide metabolism and cancer development is rapidly developing. The intervention, alteration or regulation of molecular mechanisms related to abnormal nucleotide metabolism in tumor cells has become a new idea and strategy for the treatment of tumors and prevention of recurrence and metastasis. -

Reports Report
Cell Reports Report Rhythmic Nucleotide Synthesis in the Liver: Temporal Segregation of Metabolites Jean-Michel Fustin,1 Masao Doi,1 Hiroyuki Yamada,1 Rie Komatsu,1 Shigeki Shimba,2 and Hitoshi Okamura1,* 1Department of System Biology, Graduate School of Pharmaceutical Sciences, Kyoto University, 46-29 Yoshida-Shimo-Adachi-cho, Sakyo-ku, Kyoto 606-8501, Japan 2Department of Health Science, College of Pharmacy, Nihon University, Funabashi, Chiba 274-8555, Japan *Correspondence: [email protected] DOI 10.1016/j.celrep.2012.03.001 SUMMARY (Bmal1LÀ/À), which show rhythmic activity and food intake but lack a functional molecular clock in the liver driving hepatic phys- The synthesis of nucleotides in the body is centrally iology (Shimba et al., 2011). We show that the expression of rate- controlled by the liver, via salvage or de novo limiting enzymes in nucleotide metabolism is under clock control, synthesis. We reveal a pervasive circadian influence and that nucleotides are rhythmic and time segregated. In LÀ/À on hepatic nucleotide metabolism, from rhythmic Bmal1 liver, aberrant expression of these enzymes is corre- gene expression of rate-limiting enzymes to oscil- lated with the disruption of nucleotide rhythms. In particular, lating nucleotide metabolome in wild-type (WT) ATP and was constantly low and uric acid was increased, sug- gesting inefficient purine synthesis and/or increased degrada- mice. Genetic disruption of the hepatic clock leads tion. We here provide the first (to our knowledge) integrated tran- to aberrant expression of these enzymes, together scriptome and metabolome circadian timetable focused on with anomalous nucleotide rhythms, such as a single metabolic pathway, giving insights into the physiological constant low levels of ATP with an excess in uric importance of the local hepatic clock for nucleotide synthesis. -

Nucleotide Metabolism Pathway: the Achilles' Heel for Bacterial Pathogens
REVIEW ARTICLES Nucleotide metabolism pathway: the achilles’ heel for bacterial pathogens Sujata Kumari1,2,* and Prajna Tripathi1,3 1National Institute of Immunology, New Delhi 110 067, India 2Present address: Department of Zoology, Magadh Mahila College, Patna University, Patna 800 001, India 3Present address: Institute of Molecular Medicine, Jamia Hamdard, New Delhi 110 062, India de novo pathway, the nucleotides are synthesized from Pathogens exploit their host to extract nutrients for their survival. They occupy a diverse range of host simple precursor molecules. In the salvage pathway, the niches during infection which offer variable nutrients preformed nucleobases or nucleosides which are present accessibility. To cause a successful infection a patho- in the cell or transported from external environmental gen must be able to acquire these nutrients from the milieu to the cell are utilized to form nucleotides. host as well as be able to synthesize the nutrients on its own, if required. Nucleotides are the essential me- tabolite for a pathogen and also affect the pathophysi- Purine biosynthesis pathway ology of infection. This article focuses on the role of nucleotide metabolism of pathogens during infection The purine biosynthesis pathway is universally conserved in a host. Nucleotide metabolism and disease pathoge- in living organisms (Figure 1). As an example, we here nesis are closely related in various pathogens. Nucleo- present the pathway derived from well-studied Gram- tides, purines and pyrimidines, are biosynthesized by positive bacteria Lactococcus lactis. In the de novo the de novo and salvage pathways. Whether the patho- pathway the purine nucleotides are synthesized from sim- gen will employ the de novo or salvage pathway dur- ple molecules such as phosphoribosyl pyrophosphate ing infection is dependent on various factors, like (PRPP), amino acids, CO2 and NH3 by a series of enzy- availability of nucleotides, energy condition and pres- matic reactions. -

ATP Precursor Depletion and Postischemic Myocardial Recovery'
JOURNAL OF SURGICAL RESEARCH 50,629-633 (19%) ATP Precursor Depletion and Postischemic Myocardial Recovery’ STEVENF. BOLLING, M.D.,2 EDWARDL. BOVE, M.D., AND KIM P. GALLAGHER,PH.D. Thoracic Surgery Research Laboratory, Departments of Surgery (Section of Thoracic Surgery) and Physiology, Uniuersity of Michigan Medical School, Ann Arbor, Michigan 48109 Presented at the Annual Meeting of the Association for Academic Surgery, Houston, Texas, November 14-17, 1990 lized routinely. Despite these protective techniques, how- Although cardioplegia reduces myocardial metabo- ever, adenosine triphosphate (ATP) depletion may oc- lism during ischemia, adenosine triphosphate (ATP) cur during ischemia. The slow recovery of the depleted depletion occurs, which may contribute to poor func- nucleotides, especially ATP, following ischemia is well tional recovery after reperfusion. Augmenting myo- recognized. Since intracellular ATP is essential for myo- cardial adenosine during ischemia is successful in im- cardial contraction and relaxation, ATP depletion may proving ATP repletion and myocardial recovery follow- be an important factor contributing to the incomplete ing ischemia. If adenosine is an important determinant recovery of ventricular function following ischemia and of ischemic tolerance, then depletion or elimination of reperfusion. The decrease in ATP during ischemia is not myocardial adenosine should lead to poor functional accompanied by a rise in ADP, as ADP is quickly trans- and metabolic recovery after ischemia. To test this hy- formed to AMP. The dephosphorylation of AMP by pothesis, isolated, perfused rabbit hearts were sub- 5’-nucleotidase results in formulation of adenosine jected to 120 min of 34°C ischemia. Hearts received St. which is deaminated to inosine by adenosine deaminase. -

Human Biochemistry Examination No
EXAM NUMBER:_________________ SEAT NUMBER__________________ NAME_________________________________ DABC 711 - HUMAN BIOCHEMISTRY EXAMINATION NO. 2 INSTRUCTIONS 1. Write your name on the front of the exam and on the answer sheet. 2. The exam contains 52 multiple choice question. Read each question carefully before answering. Choose the best choice from each multiple choice question and record your answer on the answer sheet using a #2 pencil. The multiple choice pquestions are worth two (2) points each. Points will not be given for a correct multiple choice answer on the exam but an incorrect answer on the answer sheet. Questions for the case study are the last questions on the exam. 3. This exam is worth 100 points (although 104 is possible). 4. Use your time wisely. All exams will be collected at 8:55 A.M. 5. GOOD LUCK ! (1) DABC 711 - HUMAN BIOCHEMISTRY February 28, 2003 1. Consider statements 1 and 2. 1. Most nucleotides used in human systems are not absorbed from the diet. 2. A nucleoside is bigger than a nucleotide. A. Statement 1 is true and Statement 2 is false. B. Statement 1 is false and Statement 2 is true. C. Both statements are true. D. Both statements are false. 2. (1) are the main precursors in the de novo synthesis of (2) . A. (1) Amino acids; (2) purines B. (1) Amino acids; (2) pyrimidines C. (1) Amino acids; (2) both purines and pyrimidines D. (1) Carbohydrates; (2) purines E. (1) Carbohydrates; (2) pyrimidines 3. PRPP is A. an enzyme used in purine synthesis. B. an enzyme used in pyrimidine synthesis. -

Development of a Genome-Scale Escherichia Coli Kinetic Metabolic Model Satisfying Flux Data for Multiple Mutant Strains
Genomic Science Contractors–Grantees Meeting XIV and USDA‐DOE Plant Feedstock Genomics for Bioenergy Meeting, March 6-9, 2016 -- http://genomicscience.energy.gov/pubs/2016abstracts/ Development of a genome-scale Escherichia coli kinetic metabolic model satisfying flux data for multiple mutant strains Ali Khodayari ([email protected]) and Costas D. Maranas Department of Chemical Engineering, Pennsylvania State University, University Park, Pennsylvania, USA. http://www.maranasgroup.com Project Goals: The goal of this effort is to construct a genome-scale kinetic model of Escherichia coli metabolism by making use of Ensemble Modeling (EM) concepts. Model parameterization is carried out using multiple flux datasets for different substrates and growth (aerobic vs. anaerobic) conditions. Kinetic modeling paradigm provides a promising platform to broaden our knowledge of cellular capacity and cell physiology beyond stoichiometric descriptions. However, developing kinetic models of metabolism at a genome-scale that faithfully recapitulate the effect of multiple genetic interventions is still an ongoing challenge. To this end, we introduce k-ecoli457, a genome-scale kinetic model of Escherichia coli metabolism that satisfies fluxomic data for a wild-type and 25 mutant strains for different substrates and growth (aerobic vs. anaerobic) conditions. The k-ecoli457 model contains 457 reactions and 337 metabolites accounting for all relevant reactions from the genome-scale iAF1260 model that carry flux under the experimental conditions of the flux measurements. These include reactions in glycolysis/gluconeogenesis, the Pentose Phosphate (PP) pathway, the TriCarboxylic Acid (TCA) cycle, anaplerotic reactions, amino acid synthesis/degradation, fatty acid oxidation/synthesis and a number of reactions in other parts of the metabolism, such as folate metabolism, cofactor and prosthetic group synthesis, alternative carbon, membrane lipid, cell envelope, nucleotide salvage and oxidative phosphorylation pathways. -
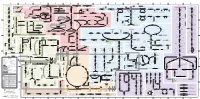
Fullsubwaymap221.Pdf
A B C D E F G H I J K L M Aldose reductase Sorbitol dehydrogenase Cholesterol synthesis Glycogen and galactose metabolism (branch point only) (debranching) glucose sorbitol fructose Aromatic amino acid metabolism Purine synthesis Pyrimidine synthesis glycogen (n+1) phenylacetate triiodothyronine (T3) dopaquinone melanin + + thyroxine (T4) (multiple steps) (many cycles) P 4' NADPH NADP NAD NADH acetyl-CoA x2 i Debranching enzyme 3' - α-1,4 bonds Aromatic amino acid 5-phosphoribosyl pyrophosphate (PRPP) HCO B 2' P 3 (branching) Glycogen 6 i (multiple steps) O H O (multiple steps) Tyrosinase O H O 1' 2 2 2 2 B 1 2 3 4 5 6 7 8 9 10 Pentose phosphate pathway Phenylalanine Tyrosine decarboxylase 6 Dopamine B Thiolase phosphorylase -5 -4 -3 -2 -1 0 1 2 3 4 Transaminase 6 glutamine 2 ATP (many cycles) Glycogen Glucose-6-phosphatase Dehydrogenase hydroxylase hydroxylase (DOPA decarboxylase) β-hydroxylase Methyltransferase 10 UDP 4:4 Transferase ATP glutathione (reduced) H O phenyllactate phenylpyruvate phenylalanine tyrosine dihydroxy- dopamine Glutamine PRPP CoA synthase Hexokinase or (in endoplasmic reticulum) 2 2 norepinephrine epinephrine glutamine Carbamoyl 9 1' phenylanine amidotransferase (GPAT) glutamate glycogen (n) Glutathione reductase Glutathione peroxidase + phosphate glycogen (n) -5 -4 -3 -2 -1 0 1 2 3 4 2' 3' 4' glucokinase 8 NADP NADPH glutamate α-ketoglutarate CO2 O H O Glycosyl (4:6) UDP-glucose ADP (L-DOPA) 2 2 synthetase II glutathione (oxidized) 2 H O α-ketoglutarate PPi glutamate acetoacetyl-CoA 7 1 Glucose -1,6-Glucosidase