Whole Genome Expression Profiling and Signal Pathway Screening of Mscs in Ankylosing Spondylitis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Active Rac1 Detection Kit 2012 09/20
Active Rac1 Detection Kit 1 Kit Orders n 877-616-CELL (2355) (30 immunoprecipitations) [email protected] Support n 877-678-TECH (8324) [email protected] Web n www.cellsignal.com rev. 09/24/20 #8815 For Research Use Only. Not For Use In Diagnostic Procedures. Species Cross-Reactivity: H, M Components Ship As: 11894S Item # Kit Quantity Storage Temp Description: The Active Rac1 Detection Kit provides all GTPgS 11521 1 X 50 µl –80°C reagents necessary for measuring activation of Rac1 GTPase in the cell. GST-PAK1-PBD fusion protein is used to bind GDP 11522 1 X 50 µl –80°C the activated form of GTP-bound Rac1, which can then be Components Ship As: 11894S Item # Kit Quantity Storage Temp immunoprecipitated with glutathione resin. Rac1 activation levels are then determined by western blot using a Rac1 GST-Human PAK1-PBD 8659 1 X 600 µg –20°C Mouse mAb. Rac1 Mouse mAb 8631 1 X 50 µl –20°C Specificity/Sensitivity: Active Rac1 Detection Kit detects Components Ship As: 11860S Item # Kit Quantity Storage Temp endogenous levels of GTP-bound (active) Rac1 as shown in Lysis/Binding/Wash Buffer 11524 1 X 100 mL 4°C Figure 1. This kit detects proteins from the indicated species, as determined through in-house testing, but may also detect Glutathione Resin 11523 1 X 3 ml 4°C homologous proteins from other species. SDS Sample Buffer 11525 1 X 1.5 ml 4°C Background: The Ras superfamily of small GTP-binding Spin Cup and Collection Tubes 11526 1 X 30 vial RT proteins (G proteins) comprise a large class of proteins (over 150 members) that can be classified into at least five families based on their sequence and functional similarities: 1 2 3 4 Figure 1. -

Rac1 Accumulates in the Nucleus During the G2 Phase of the Cell
Published Online: 28 April, 2008 | Supp Info: http://doi.org/10.1083/jcb.200801047 JCB: ARTICLE Downloaded from jcb.rupress.org on May 9, 2018 Rac1 accumulates in the nucleus during the G2 phase of the cell cycle and promotes cell division David Michaelson , 2,5 Wasif Abidi , 2,5 Daniele Guardavaccaro , 4,5 Mo Zhou, 3,5 Ian Ahearn , 3,5 Michele Pagano , 4,5 and Mark R. Philips 1,2,3,5 1 Department of Medicine, 2 Department of Cell Biology, 3 Department of Pharmacology, 4 Department of Pathology, and the 5 New York University Cancer Institute, New York University School of Medicine, New York, NY 10016 ac1 regulates a wide variety of cellular processes. zation of cells stably expressing low levels of GFP-Rac1, The polybasic region of the Rac1 C terminus func- and time-lapse microscopy of asynchronous cells revealed Rtions both as a plasma membrane – targeting motif Rac1 accumulation in the nucleus in late G2 and exclu- and a nuclear localization sequence (NLS). We show that sion in early G1. Although constitutively active Rac1 re- a triproline N-terminal to the polybasic region contributes stricted to the cytoplasm inhibited cell division, activated to the NLS, which is cryptic in the sense that it is strongly Rac1 expressed constitutively in the nucleus increased the inhibited by geranylgeranylation of the adjacent cysteine. mitotic rate. These results show that Rac1 cycles in and out Subcellular fractionation demonstrated endogenous Rac1 of the nucleus during the cell cycle and thereby plays a in the nucleus and Triton X-114 partition revealed that this role in promoting cell division. -

Role of Rac1-Pak Pathway in Aggressive B-Cell Lymphoma
University of Nebraska Medical Center DigitalCommons@UNMC Theses & Dissertations Graduate Studies Spring 5-4-2019 Role of Rac1-Pak pathway in aggressive b-cell lymphoma Tian Tian University of Nebraska Medical Center Follow this and additional works at: https://digitalcommons.unmc.edu/etd Part of the Hemic and Lymphatic Diseases Commons, and the Neoplasms Commons Recommended Citation Tian, Tian, "Role of Rac1-Pak pathway in aggressive b-cell lymphoma" (2019). Theses & Dissertations. 344. https://digitalcommons.unmc.edu/etd/344 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@UNMC. It has been accepted for inclusion in Theses & Dissertations by an authorized administrator of DigitalCommons@UNMC. For more information, please contact [email protected]. i Role of Rac1-PAK Pathway in Aggressive B-cell Lymphomas By Tian Tian A DISSERTATION Presented to the Faculty of The University of Nebraska Graduate College in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Pathology & Microbiology Graduate program Under the Supervision of Professor Kai Fu University of Nebraska Medical Center, Omaha, Nebraska Dec, 2018 Supervisory Committee: Ying Yan, Ph.D. John S Davis, Ph.D. Timothy C Greiner, M.D. Javeed Iqbal, Ph.D. ii Role of Rac1-PAK pathway in Aggressive B-cell Lymphomas Tian Tian University of Nebraska Medical Center, 2018 Advisor: Kai Fu, M.D. Ph.D. Aggressive B-cell lymphomas are diverse group of neoplasms that arise at different stages of B-cell development and by various mechanisms of neoplastic transformation. Aggressive B-cell lymphomas include many types, subtypes and variants of diffuse large B-cell lymphoma (DLBCL), Burkitt lymphoma (BL), mantle cell lymphoma (MCL) and B lymphoblastic lymphoma. -

Rac1 Promotes Cell Motility by Controlling Cell Mechanics in Human Glioblastoma
cancers Article Rac1 Promotes Cell Motility by Controlling Cell Mechanics in Human Glioblastoma Jing Xu 1,2, Nicola Galvanetto 1, Jihua Nie 1,3, Yili Yang 2,* and Vincent Torre 1,2,3,4,* 1 International School for Advanced Studies (SISSA), 34136 Trieste, Italy; [email protected] (J.X.); [email protected] (N.G.); [email protected] (J.N.) 2 Joint Laboratory of Biophysics and Translational Medicine, Suzhou Institute of Systems Medicine (ISM)- International School for Advanced Studies (SISSA), Suzhou 215123, China 3 School of Radiation Medicine and Protection, State Key Laboratory of Radiation Medicine and Protection Medical College of Soochow University, Suzhou 215123, China 4 Cixi Institute of Biomedical Engineering, Ningbo Institute of Materials Technology and Engineering Chinese Academy of Sciences, Ningbo 315201, China * Correspondence: [email protected] (Y.Y.);[email protected] (V.T.) Received: 21 May 2020; Accepted: 2 June 2020; Published: 23 June 2020 Abstract: The failure of existing therapies in treating human glioblastoma (GBM) mostly is due to the ability of GBM to infiltrate into healthy regions of the brain; however, the relationship between cell motility and cell mechanics is not well understood. Here, we used atomic force microscopy (AFM), live-cell imaging, and biochemical tools to study the connection between motility and mechanics in human GBM cells. It was found thatRac1 inactivation by genomic silencing and inhibition with EHT 1864 reduced cell motility, inhibited cell ruffles, and disrupted the dynamics of cytoskeleton organization and cell adhesion. These changes were correlated with abnormal localization of myosin IIa and a rapid suppression of the phosphorylation of Erk1/2. -

G Protein Regulation of MAPK Networks
Oncogene (2007) 26, 3122–3142 & 2007 Nature Publishing Group All rights reserved 0950-9232/07 $30.00 www.nature.com/onc REVIEW G Protein regulation of MAPK networks ZG Goldsmith and DN Dhanasekaran Fels Institute for Cancer Research and Molecular Biology, Temple University School of Medicine, Philadelphia, PA, USA G proteins provide signal-coupling mechanisms to hepta- the a-subunits has been used as a basis for the helical cell surface receptors and are criticallyinvolved classification of G proteins into Gs,Gi,Gq and G12 in the regulation of different mitogen-activated protein families in which the a-subunits that show more than kinase (MAPK) networks. The four classes of G proteins, 50% homology are grouped together (Simon et al., defined bythe G s,Gi,Gq and G12 families, regulate 1991). In G-protein-coupled receptor (GPCR)-mediated ERK1/2, JNK, p38MAPK, ERK5 and ERK6 modules by signaling pathways, ligand-activated receptors catalyse different mechanisms. The a- as well as bc-subunits are the exchange of the bound GDP to GTP in the a-subunit involved in the regulation of these MAPK modules in a following which the GTP-bound a-subunit disassociate context-specific manner. While the a- and bc-subunits from the receptor as well as the bg-subunit. The GTP- primarilyregulate the MAPK pathwaysvia their respec- bound a-subunit and the bg-subunit stimulate distinct tive effector-mediated signaling pathways, recent studies downstream effectors including enzymes, ion channels have unraveled several novel signaling intermediates and small GTPase, thus regulating multiple signaling including receptor tyrosine kinases and small GTPases pathways including those involved in the activation of through which these G-protein subunits positivelyas well mitogen-activated protein kinase (MAPK) modules as negativelyregulate specific MAPK modules. -

A Link Between Planar Polarity and Staircase-Like Bundle Architecture in Hair Cells Basile Tarchini1,2,3,4,*, Abigail L
© 2016. Published by The Company of Biologists Ltd | Development (2016) 143, 3926-3932 doi:10.1242/dev.139089 RESEARCH REPORT A link between planar polarity and staircase-like bundle architecture in hair cells Basile Tarchini1,2,3,4,*, Abigail L. D. Tadenev1, Nicholas Devanney1 and Michel Cayouette4,5,6,* ABSTRACT However, the mechanisms that instruct morphological and Sensory perception in the inner ear relies on the hair bundle, the highly molecular asymmetry across the bundle remain unknown. polarized brush of movement detectors that crowns hair cells. We External cues, including a role for the extracellular matrix or the previously showed that, in the mouse cochlea, the edge of the forming kinocilium, have been proposed but not experimentally validated bundle is defined by the ‘bare zone’, a microvilli-free sub-region of (Manor and Kachar, 2008). apical membrane specified by the Insc-LGN-Gαi protein complex. We Tissue and cell polarization during early development lay the now report that LGN and Gαi also occupy the very tip of stereocilia that structural foundations required for sensory function. Before a directly abut the bare zone. We demonstrate that LGN and Gαi are both marked staircase pattern and tip-links arise postnatally in the mouse ‘ ’ essential for promoting the elongation and differential identity of cochlea, core planar cell polarity (PCP) proteins that are stereocilia across rows. Interestingly, we also reveal that total LGN-Gαi asymmetrically enriched at cell-cell junctions ensure uniform protein amounts are actively balanced between the bare zone and bundle orientation across HCs (May-Simera and Kelley, 2012). stereocilia tips, suggesting that early planar asymmetry of protein Furthermore, recent studies have clarified that planar asymmetry of enrichment at the bare zone confers adjacent stereocilia their tallest the cytoskeleton in each HC (which is manifested, for example, by identity. -

A Rhog-Mediated Signaling Pathway That Modulates Invadopodia Dynamics in Breast Cancer Cells Silvia M
© 2017. Published by The Company of Biologists Ltd | Journal of Cell Science (2017) 130, 1064-1077 doi:10.1242/jcs.195552 RESEARCH ARTICLE A RhoG-mediated signaling pathway that modulates invadopodia dynamics in breast cancer cells Silvia M. Goicoechea, Ashtyn Zinn, Sahezeel S. Awadia, Kyle Snyder and Rafael Garcia-Mata* ABSTRACT micropinocytosis, bacterial uptake, phagocytosis and leukocyte One of the hallmarks of cancer is the ability of tumor cells to invade trans-endothelial migration (deBakker et al., 2004; Ellerbroek et al., surrounding tissues and metastasize. During metastasis, cancer cells 2004; Jackson et al., 2015; Katoh et al., 2006, 2000; van Buul et al., degrade the extracellular matrix, which acts as a physical barrier, by 2007). Recent studies have revealed that RhoG plays a role in tumor developing specialized actin-rich membrane protrusion structures cell invasion and may contribute to the formation of invadopodia called invadopodia. The formation of invadopodia is regulated by Rho (Hiramoto-Yamaki et al., 2010; Kwiatkowska et al., 2012). GTPases, a family of proteins that regulates the actin cytoskeleton. Invadopodia are actin-rich adhesive structures that form in the Here, we describe a novel role for RhoG in the regulation of ventral surface of cancer cells and allow them to degrade the invadopodia disassembly in human breast cancer cells. Our results extracellular matrix (ECM) (Gimona et al., 2008). Formation of show that RhoG and Rac1 have independent and opposite roles invadopodia involves a series of steps that include the disassembly in the regulation of invadopodia dynamics. We also show that SGEF of focal adhesions and stress fibers, and the relocalization of several (also known as ARHGEF26) is the exchange factor responsible of their components into the newly formed invadopodia (Hoshino for the activation of RhoG during invadopodia disassembly. -

Screening of Potential Genes and Transcription Factors Of
ANIMAL STUDY e-ISSN 1643-3750 © Med Sci Monit, 2018; 24: 503-510 DOI: 10.12659/MSM.907445 Received: 2017.10.08 Accepted: 2018.01.01 Screening of Potential Genes and Transcription Published: 2018.01.25 Factors of Postoperative Cognitive Dysfunction via Bioinformatics Methods Authors’ Contribution: ABE 1 Yafeng Wang 1 Department of Anesthesiology, The First Affiliated Hospital of Guangxi Medical Study Design A AB 1 Ailan Huang University, Nanning, Guangxi, P.R. China Data Collection B 2 Department of Gynecology, People’s Hospital of Guangxi Zhuang Autonomous Statistical Analysis C BEF 1 Lixia Gan Region, The First Affiliated Hospital of Guangxi Medical University, Nanning, Data Interpretation D BCF 1 Yanli Bao Guangxi, P.R. China Manuscript Preparation E BDF 1 Weilin Zhu 3 Department of Anesthesiology, The First Affiliated Hospital of Guangxi Medical Literature Search F University, Nanning, Guangxi, P.R. China Funds Collection G EF 1 Yanyan Hu AE 1 Li Ma CF 2 Shiyang Wei DE 3 Yuyan Lan Corresponding Author: Yafeng Wang, e-mail: [email protected] Source of support: Departmental sources Background: The aim of this study was to explore the potential genes and transcription factors involved in postoperative cognitive dysfunction (POCD) via bioinformatics analysis. Material/Methods: GSE95070 miRNA expression profiles were downloaded from Gene Expression Omnibus database, which in- cluded five hippocampal tissues from POCD mice and controls. Moreover, the differentially expressed miRNAs (DEMs) between the two groups were identified. In addition, the target genes of DEMs were predicted using Targetscan 7.1, followed by protein-protein interaction (PPI) network construction, functional enrichment anal- ysis, pathway analysis, and prediction of transcription factors (TFs) targeting the potential targets. -
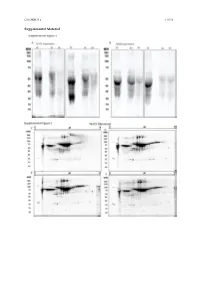
Type of the Paper (Article
Cells 2020, 9, x 1 of 19 Supplemental Material Cells 2020, 9, x 2 of 19 Figure S1. Secretome enrichment: protocol optimization. 1D SDS-PAGE documentation of washing steps: Culture medium was substituted with FCS-free medium, which was changed every 2 h. The supernatants were then collected, and the proteins isolated and separated in 1D SDS-PAGE ((A) TK173 and (B) TK188). Proteins were stained with Flamingo fluorescent gel stain. Two-dimensional pattern of the proteins isolated from supernatant of TK173, (C) 2 h, (D) 4 h, (E) 6 h, and (F) 8 h after changing to FCS-free medium. (G) Cell secretome collected 24 h after elimination of the contaminating FCS-proteins with different washing steps. Proteins were stained with Flamingo fluorescent gel stain. Cells 2020, 9, x 3 of 19 Figure S2. 2-DE reference maps of secretomes; 150 μg proteins were loaded on an 11 cm IPG strip with a linear pH gradient PI 5–8 for IEF; 12% SDS-polyacrylamide gels were used for the second dimension. Proteins were stained with Flamingo fluorescent gel stain. Identified spots were assigned a number corresponding to that in their table. 2-DE maps from secretome of (A) TK173 control and (B) TGFβ1- treated ones. The 2-DE patterns revealed an alteration of secretome in stimulated TK173. Secretome patterns from TK173 treated with (C) ANG II and (D) PDGF. Cells 2020, 9, x 4 of 19 A Figure S3. Classification of the differentially expressed proteins upon ANG II, TGFβ1, or PDGF treatment in TK173. (A) Bar charts of the cellular component analyzed by STRAP biological function analysis in which the identified proteins from all treatments in both cell types are involved. -

Critical Activities of Rac1 and Cdc42hs in Skeletal Myogenesis
Critical Activities of Rac1 and Cdc42Hs in Skeletal Myogenesis: Antagonistic Effects of JNK and p38 Pathways Mayya Meriane, Pierre Roux, Michael Primig, Philippe Fort, Cécile Gauthier-Rouvière To cite this version: Mayya Meriane, Pierre Roux, Michael Primig, Philippe Fort, Cécile Gauthier-Rouvière. Critical Activities of Rac1 and Cdc42Hs in Skeletal Myogenesis: Antagonistic Effects of JNK and p38 Path- ways. Molecular Biology of the Cell, American Society for Cell Biology, 2000, 11 (8), pp.2513-2528. 10.1091/mbc.11.8.2513. hal-02267479 HAL Id: hal-02267479 https://hal.archives-ouvertes.fr/hal-02267479 Submitted on 26 Jun 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - ShareAlike| 4.0 International License Molecular Biology of the Cell Vol. 11, 2513–2528, August 2000 Critical Activities of Rac1 and Cdc42Hs in Skeletal Myogenesis: Antagonistic Effects of JNK and p38 Pathways Mayya Meriane,* Pierre Roux,* Michael Primig,† Philippe Fort,* and Ce´cile Gauthier-Rouvie`re*‡ *Centre de Recherche de Biochimie Macromole´culaire, Centre National de la Recherche Scientifique Unite´ Propre de Recherche 1086, 34293 Montpellier Cedex, France; and †Institut de Ge´ne´tique Humaine, Centre National de la Recherche Scientifique Unite´ Propre de Recherche 1142, 34396 Montpellier Cedex 5, France Submitted January 5, 2000; Revised April 7, 2000; Accepted May 19, 2000 Monitoring Editor: Paul T. -

Beyond Traditional Morphological Characterization of Lung
Cancers 2020 S1 of S15 Beyond Traditional Morphological Characterization of Lung Neuroendocrine Neoplasms: In Silico Study of Next-Generation Sequencing Mutations Analysis across the Four World Health Organization Defined Groups Giovanni Centonze, Davide Biganzoli, Natalie Prinzi, Sara Pusceddu, Alessandro Mangogna, Elena Tamborini, Federica Perrone, Adele Busico, Vincenzo Lagano, Laura Cattaneo, Gabriella Sozzi, Luca Roz, Elia Biganzoli and Massimo Milione Table S1. Genes Frequently mutated in Typical Carcinoids (TCs). Mutation Original Entrez Gene Gene Rate % eukaryotic translation initiation factor 1A X-linked [Source: HGNC 4.84 EIF1AX 1964 EIF1AX Symbol; Acc: HGNC: 3250] AT-rich interaction domain 1A [Source: HGNC Symbol;Acc: HGNC: 4.71 ARID1A 8289 ARID1A 11110] LDL receptor related protein 1B [Source: HGNC Symbol; Acc: 4.35 LRP1B 53353 LRP1B HGNC: 6693] 3.53 NF1 4763 NF1 neurofibromin 1 [Source: HGNC Symbol;Acc: HGNC: 7765] DS cell adhesion molecule like 1 [Source: HGNC Symbol; Acc: 2.90 DSCAML1 57453 DSCAML1 HGNC: 14656] 2.90 DST 667 DST dystonin [Source: HGNC Symbol;Acc: HGNC: 1090] FA complementation group D2 [Source: HGNC Symbol; Acc: 2.90 FANCD2 2177 FANCD2 HGNC: 3585] piccolo presynaptic cytomatrix protein [Source: HGNC Symbol; Acc: 2.90 PCLO 27445 PCLO HGNC: 13406] erb-b2 receptor tyrosine kinase 2 [Source: HGNC Symbol; Acc: 2.44 ERBB2 2064 ERBB2 HGNC: 3430] BRCA1 associated protein 1 [Source: HGNC Symbol; Acc: HGNC: 2.35 BAP1 8314 BAP1 950] capicua transcriptional repressor [Source: HGNC Symbol; Acc: 2.35 CIC 23152 CIC HGNC: -

New Structural Perspectives in G Protein-Coupled Receptor-Mediated Src Family Kinase Activation
International Journal of Molecular Sciences Review New Structural Perspectives in G Protein-Coupled Receptor-Mediated Src Family Kinase Activation Sandra Berndt * and Ines Liebscher Rudolf Schönheimer Institute of Biochemistry, Molecular Biochemistry, Medical Faculty, University of Leipzig, 04103 Leipzig, Germany; [email protected] * Correspondence: [email protected]; Tel.: +49-341-9722175 Abstract: Src family kinases (SFKs) are key regulators of cell proliferation, differentiation, and survival. The expression of these non-receptor tyrosine kinases is strongly correlated with cancer development and tumor progression. Thus, this family of proteins serves as an attractive drug target. The activation of SFKs can occur via multiple signaling pathways, yet many of them are poorly understood. Here, we summarize the current knowledge on G protein-coupled receptor (GPCR)- mediated regulation of SFKs, which is of considerable interest because GPCRs are among the most widely used pharmaceutical targets. This type of activation can occur through a direct interaction between the two proteins or be allosterically regulated by arrestins and G proteins. We postulate that a rearrangement of binding motifs within the active conformation of arrestin-3 mediates Src regulation by comparison of available crystal structures. Therefore, we hypothesize a potentially different activation mechanism compared to arrestin-2. Furthermore, we discuss the probable direct regulation of SFK by GPCRs and investigate the intracellular domains of exemplary GPCRs with conserved polyproline binding motifs that might serve as scaffolding domains to allow such a direct interaction. Large intracellular domains in GPCRs are often understudied and, in general, not much Citation: Berndt, S.; Liebscher, I. is known of their contribution to different signaling pathways.