Combining Faceted Navigation and Research Group Detection for COVID-19 Exploratory Scientific Search
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Critical Thinking for Language Models
Critical Thinking for Language Models Gregor Betz Christian Voigt Kyle Richardson KIT KIT Allen Institute for AI Karlsruhe, Germany Karlsruhe, Germany Seattle, WA, USA [email protected] [email protected] [email protected] Abstract quality of online debates (Hansson, 2004; Guia¸su and Tindale, 2018; Cheng et al., 2017): Neural lan- This paper takes a first step towards a critical guage models are known to pick up and reproduce thinking curriculum for neural auto-regressive normative biases (e.g., regarding gender or race) language models. We introduce a synthetic present in the dataset they are trained on (Gilburt corpus of deductively valid arguments, and generate artificial argumentative texts to train and Claydon, 2019; Blodgett et al., 2020; Nadeem CRiPT: a critical thinking intermediarily pre- et al., 2020), as well as other annotation artifacts trained transformer based on GPT-2. Signifi- (Gururangan et al., 2018); no wonder this happens cant transfer learning effects can be observed: with argumentative biases and reasoning flaws, too Trained on three simple core schemes, CRiPT (Kassner and Schütze, 2020; Talmor et al., 2020). accurately completes conclusions of different, This diagnosis suggests that there is an obvious and more complex types of arguments, too. remedy for LMs’ poor reasoning capability: make CRiPT generalizes the core argument schemes in a correct way. Moreover, we obtain con- sure that the training corpus contains a sufficient sistent and promising results for NLU bench- amount of exemplary episodes of sound reasoning. marks. In particular, CRiPT’s zero-shot accu- In this paper, we take a first step towards the cre- racy on the GLUE diagnostics exceeds GPT- ation of a “critical thinking curriculum” for neural 2’s performance by 15 percentage points. -

Can We Make Text-Based AI Less Racist, Please?
Can We Make Text-Based AI Less Racist, Please? Last summer, OpenAI launched GPT-3, a state-of-the-art artificial intelligence contextual language model that promised computers would soon be able to write poetry, news articles, and programming code. Sadly, it was quickly found to be foulmouthed and toxic. OpenAI researchers say they’ve found a fix to curtail GPT-3’s toxic text by feeding the programme roughly a hundred encyclopedia-like samples of writing on usual topics like history and technology, but also extended topics such as abuse, violence and injustice. You might also like: Workforce shortages are an issue faced by many hospital leaders. Implementing four basic child care policies in your institution, could be a game-changer when it comes to retaining workers - especially women, according to a recent article published in Harvard Business Review (HBR). Learn more GPT-3 has shown impressive ability to understand and compose language. It can answer SAT analogy questions better than most people, and it was able to fool community forum members online. More services utilising these large language models, which can interpret or generate text, are being offered by big tech companies everyday. Microsoft is using GPT-3 in its' programming and Google considers these language models to be crucial in the future of search engines. OpenAI’s project shows how a technology that has shown enormous potential can also spread disinformation and perpetuate biases. Creators of GPT-3 knew early on about its tendency to generate racism and sexism. OpenAI released a paper in May 2020, before GPT-3 was licensed to developers. -

Semantic Scholar Adds 25 Million Scientific Papers in 2020 Through New Publisher Partnerships
Press Release Semantic Scholar Adds 25 Million Scientific Papers in 2020 Through New Publisher Partnerships Cambridge University Press, Wiley, and the University of Chicago Press are the latest publishers to partner with Semantic Scholar to expand discovery of scientific research Seattle, WA | December 14, 2020 Researchers and academics around the world can now discover academic literature from leading publishers including Cambridge University Press, Wiley, and The University of Chicago Press using Semantic Scholar, a free AI-powered research tool for academic papers from the Allen Institute for AI. “We are thrilled to have grown our corpus by more than 25 million papers this year, thanks to our new partnerships with top academic publishers,” says Sebastian Kohlmeier, head of partnerships and operations for Semantic Scholar at AI2. “By adding hundreds of peer-reviewed journals to our corpus we’re able to better serve the needs of researchers everywhere.” Semantic Scholar’s millions of users can now use innovative AI-powered features to explore peer-reviewed research from these extensive journal collections, covering all academic disciplines. Cambridge University Press is part of the University of Cambridge and publishes a wide range of academic content in all fields of study. It has provided more than 380 peer-reviewed journals in subjects ranging from astronomy to the arts, mathematics, and social sciences to Semantic Scholar’s corpus. Peter White, the Press’s Manager for Digital Partnerships, said: “The academic communities we serve increasingly engage with research online, a trend which has been further accelerated by the pandemic. We are confident this agreement with Semantic Scholar will further enhance the discoverability of our content, helping researchers to find what they need faster and increasing the reach, use and impact of the research we publish.” Wiley is an innovative, global publishing leader and has been a trusted source of scientific content for more than 200 years. -

AI Watch Artificial Intelligence in Medicine and Healthcare: Applications, Availability and Societal Impact
JRC SCIENCE FOR POLICY REPORT AI Watch Artificial Intelligence in Medicine and Healthcare: applications, availability and societal impact EUR 30197 EN This publication is a Science for Policy report by the Joint Research Centre (JRC), the European Commission’s science and knowledge service. It aims to provide evidence-based scientific support to the European policymaking process. The scientific output expressed does not imply a policy position of the European Commission. Neither the European Commission nor any person acting on behalf of the Commission is responsible for the use that might be made of this publication. For information on the methodology and quality underlying the data used in this publication for which the source is neither Eurostat nor other Commission services, users should contact the referenced source. The designations employed and the presentation of material on the maps do not imply the expression of any opinion whatsoever on the part of the European Union concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Contact information Email: [email protected] EU Science Hub https://ec.europa.eu/jrc JRC120214 EUR 30197 EN PDF ISBN 978-92-76-18454-6 ISSN 1831-9424 doi:10.2760/047666 Luxembourg: Publications Office of the European Union, 2020. © European Union, 2020 The reuse policy of the European Commission is implemented by the Commission Decision 2011/833/EU of 12 December 2011 on the reuse of Commission documents (OJ L 330, 14.12.2011, p. 39). Except otherwise noted, the reuse of this document is authorised under the Creative Commons Attribution 4.0 International (CC BY 4.0) licence (https://creativecommons.org/licenses/by/4.0/). -

AI Can Recognize Images. but Can It Understand This Headline?
10/18/2019 AI Can Recognize Images, But Text Has Been Tricky—Until Now | WIRED SUBSCRIBE GREGORY BARBER B U S I N E S S 09.07.2018 01:55 PM AI Can Recognize Images. But Can It Understand This Headline? New approaches foster hope that computers can comprehend paragraphs, classify email as spam, or generate a satisfying end to a short story. CASEY CHIN 3 FREE ARTICLES LEFT THIS MONTH Get unlimited access. Subscribe https://www.wired.com/story/ai-can-recognize-images-but-understand-headline/ 1/9 10/18/2019 AI Can Recognize Images, But Text Has Been Tricky—Until Now | WIRED In 2012, artificial intelligence researchers revealed a big improvement in computers’ ability to recognize images by SUBSCRIBE feeding a neural network millions of labeled images from a database called ImageNet. It ushered in an exciting phase for computer vision, as it became clear that a model trained using ImageNet could help tackle all sorts of image- recognition problems. Six years later, that’s helped pave the way for self-driving cars to navigate city streets and Facebook to automatically tag people in your photos. In other arenas of AI research, like understanding language, similar models have proved elusive. But recent research from fast.ai, OpenAI, and the Allen Institute for AI suggests a potential breakthrough, with more robust language models that can help researchers tackle a range of unsolved problems. Sebastian Ruder, a researcher behind one of the new models, calls it his field’s “ImageNet moment.” The improvements can be dramatic. The most widely tested model, so far, is called Embeddings from Language Models, or ELMo. -

AI Ignition Ignite Your AI Curiosity with Oren Etzioni AI for the Common Good
AI Ignition Ignite your AI curiosity with Oren Etzioni AI for the common good From Deloitte’s AI Institute, this is AI Ignition—a monthly chat about the human side of artificial intelligence with your host, Beena Ammanath. We will take a deep dive into the past, present, and future of AI, machine learning, neural networks, and other cutting-edge technologies. Here is your host, Beena. Beena Ammanath (Beena): Hello, my name is Beena Ammanath. I am the executive director of the Deloitte AI Institute, and today on AI Ignition we have Oren Etzioni, a professor, an entrepreneur, and the CEO of Allen Institute for AI. Oren has helped pioneer meta search, online comparison shopping, machine reading, and open information extraction. He was also named Seattle’s Geek of the Year in 2013. Welcome to the show, Oren. I am so excited to have you on our AI Ignition show today. How are you doing? Oren Etzioni (Oren): I am doing as well as anyone can be doing in these times of COVID and counting down the days till I get a vaccine. Beena: Yeah, and how have you been keeping yourself busy during the pandemic? Oren: Well, we are fortunate that AI is basically software with a little sideline into robotics, and so despite working from home, we have been able to stay productive and engaged. It’s just a little bit less fun because one of the things I like to say about AI is that it’s still 99% human intelligence, and so obviously the human interactions are stilted. -
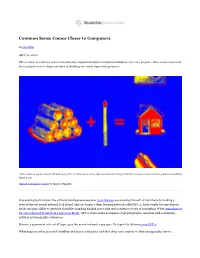
Common Sense Comes Closer to Computers
Common Sense Comes Closer to Computers By John Pavlus April 30, 2020 The problem of common-sense reasoning has plagued the field of artificial intelligence for over 50 years. Now a new approach, borrowing from two disparate lines of thinking, has made important progress. Add a match to a pile of wood. What do you get? For a human, it’s easy. But machines have long lacked the common-sense reasoning abilities needed to figure it out. Guillem Casasús Xercavins for Quanta Magazine One evening last October, the artificial intelligence researcher Gary Marcus was amusing himself on his iPhone by making a state-of-the-art neural network look stupid. Marcus’ target, a deep learning network called GPT-2, had recently become famous for its uncanny ability to generate plausible-sounding English prose with just a sentence or two of prompting. When journalists at The Guardian fed it text from a report on Brexit, GPT-2 wrote entire newspaper-style paragraphs, complete with convincing political and geographic references. Marcus, a prominent critic of AI hype, gave the neural network a pop quiz. He typed the following into GPT-2: What happens when you stack kindling and logs in a fireplace and then drop some matches is that you typically start a … Surely a system smart enough to contribute to The New Yorker would have no trouble completing the sentence with the obvious word, “fire.” GPT-2 responded with “ick.” In another attempt, it suggested that dropping matches on logs in a fireplace would start an “irc channel full of people.” Marcus wasn’t surprised. -

Oren Etzioni, Phd: CEO of Allen Institute for AI
Behind the Tech Kevin Scott Podcast EP-26: Oren Etzioni, PhD: CEO of Allen Institute for AI [MUSIC] OREN ETZIONI: Whose responsibility is it? The responsibility and liability has to ultimately rest with a person. You can’t say, “Hey, you know, look, my car ran you over, it’s an AI car, I don’t know what it did, it’s not my fault, right?” You as the driver or maybe it’s the manufacturer if there’s some malfunction, but people have to be responsible for the behavior of the machines. [MUSIC] KEVIN SCOTT: Hi, everyone. Welcome to Behind the Tech. I'm your host, Kevin Scott, Chief Technology Officer for Microsoft. In this podcast, we're going to get behind the tech. We'll talk with some of the people who have made our modern tech world possible and understand what motivated them to create what they did. So, join me to maybe learn a little bit about the history of computing and get a few behind- the-scenes insights into what's happening today. Stick around. [MUSIC] CHRISTINA WARREN: Hello, and welcome to Behind the Tech. I’m Christina Warren, senior cloud advocate at Microsoft. KEVIN SCOTT: And I’m Kevin Scott. CHRISTINA WARREN: Today on the show, our guest is Oren Etzioni. Oren is a professor, entrepreneur, and is the chief executive officer for the Allen Institute for AI. So, Kevin, I’m guessing that you guys have already crossed paths in your professional pursuits. KEVIN SCOTT: Yeah, I’ve been lucky enough to know Oren for the past several years. -

Artificial Intelligence's Grand Challenges
Article Artificial Intelligence’s Grand Challenges: Past, Present, and Future Ganesh Mani Innovative, bold initiatives that cap- rand challenges are important as they act as compasses ture the imagination of researchers and for researchers and practitioners alike — especially system builders are often required to young professionals — who are pondering worth- spur a field of science or technology for- G while problems to work on, testing the boundaries of what ward. A vision for the future of artifi- is possible! Challenge tasks also unleash the competitive cial intelligence was laid out by Turing spirit in participants as evidenced by the plethora of active Award winner Raj Reddy in his 1988 Presidential address to the Associa- participants in Kaggle competitions (and forum discussions tion for the Advancement of Artificial therein). Prize money and research bragging rights also Intelligence. It is time to provide an accrue to the winners. The Defense Advanced Research Pro- accounting of the progress that has jects Agency Grand Challenges1 and X prizes2 are some of been made in the field, over the last the best-known successful programs that have helped make three decades, toward the challenge significant progress across many domains applying artifi- goals. While some tasks such as the cial intelligence (AI). As grand challenges are accomplished, world-champion chess machine were other than the long-term benefits the solutions engender, accomplished in short order, many the positive press they garner helps rally society behind others, such as self-replicating sys- the field. Trickle-down benefits include renewed respect for tems, require more focus and break- throughs for completion. -

Commonsenseqa 2.0: Exposing the Limits of AI Through Gamification
CommonsenseQA 2.0: Exposing the Limits of AI through Gamification Alon Talmor1 Ori Yoran1;2 Ronan Le Bras1 Chandra Bhagavatula1 Yoav Goldberg1 Yejin Choi1;3 Jonathan Berant1;2 1The Allen Institute for AI, 2Tel-Aviv University, 3University of Washington {ronanlb,chandrab,yoavg,yejinc,jonathan}@allenai.org {alontalmor,oriy}@mail.tau.ac.il Abstract 1 Constructing benchmarks that test the abilities of modern natural language un- 2 derstanding models is difficult – pre-trained language models exploit artifacts in 3 benchmarks to achieve human parity, but still fail on adversarial examples and make 4 errors that demonstrate a lack of common sense. In this work, we propose gamifi- 5 cation as a framework for data construction. The goal of players in the game is to 6 compose questions that mislead a rival AI, while using specific phrases for extra 7 points. The game environment leads to enhanced user engagement and simultane- 8 ously gives the game designer control over the collected data, allowing us to collect 9 high-quality data at scale. Using our method we create CommonsenseQA 2.0, 10 which includes 14,343 yes/no questions, and demonstrate its difficulty for models 11 that are orders-of-magnitude larger than the AI used in the game itself. Our best 12 baseline, the T5-based UNICORN with 11B parameters achieves an accuracy of 13 70:2%, substantially higher than GPT-3 (52:9%) in a few-shot inference setup. 14 Both score well below human performance which is at 94:1%. 15 1 Introduction 16 Evaluating models for natural language understanding (NLU) has become a frustrating endeavour. -

Artificial Intelligence
Artificial Intelligence A White Paper Jonathan Kirk, Co-Founder & Director of AI Strategy systemsGo www.systemsGo.asia January 2018 Table of Contents Big picture ..................................................................................................................... 4 What is AI anyway? ....................................................................................................... 5 Why now? What's changed? ........................................................................................ 5 Old algorithms, new life ................................................................................................ 6 Data - big and getting bigger ........................................................................................ 8 Some key data initiatives ......................................................................................................8 Computing power: continuing to explode .................................................................. 9 The state of AI today: better than some humans ....................................................... 9 AI to AGI ....................................................................................................................... 10 AI at work ..................................................................................................................... 12 Some examples of AI in use today - by industry...............................................................12 Emerging applications ........................................................................................................13 -

Toward Automatic Bootstrapping of Online Communities Using
Toward Automatic Bootstrapping of Online Communities Using Decision-theoretic Optimization Shih-Wen Huang* Jonathan Bragg* Isaac Cowhey University of Washington University of Washington Allen Institute for AI Seattle, WA, USA Seattle, WA, USA Seattle, WA, USA [email protected] [email protected] [email protected] Oren Etzioni Daniel S. Weld Allen Institute for AI University of Washington Seattle, WA, USA Seattle, WA, USA [email protected] [email protected] ABSTRACT INTRODUCTION Successful online communities (e.g., Wikipedia, Yelp, and The Internet has spawned communities that create extraordi- StackOverflow) can produce valuable content. However, nary resources. For example, Wikipedia’s 23 million users many communities fail in their initial stages. Starting an on- have created over 4 million English articles, a resource over line community is challenging because there is not enough 100 times larger than any other encyclopedia. Similarly, content to attract a critical mass of active members. This StackOverflow has become a top resource for programmers paper examines methods for addressing this cold-start prob- with 14 million answers to 8.5 million questions, while Yelp lem in datamining-bootstrappable communities by attract- users generated more than 67 million reviews.1 ing non-members to contribute to the community. We make four contributions: 1) we characterize a set of communi- In reality, however, most online communities fail. For exam- ple, thousands of open source projects have been created on ties that are “datamining-bootstrappable” and define the boot- SourceForge, but only 10% have three or more members [24]. strapping problem in terms of decision-theoretic optimiza- Furthermore, more than 50% of email-based groups received tion, 2) we estimate the model parameters in a case study involving the Open AI Resources website, 3) we demonstrate no messages during a four-month study period [6].