Speciation, Evolution and Phylogeny of Some Shallow-Water Octocorals (Cnidaria: Anthozoa)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Di Camillo Et Al 2017
This is a post-peer-review, pre-copyedit version of an article published in Biodiversity and Conservation on 23 December 2017 (First Online). The final authenticated version is available online at: https://doi.org/10.1007/s10531-017-1492-8 https://link.springer.com/article/10.1007%2Fs10531-017-1492-8 An embargo period of 12 months applies to this Journal. This paper has received funding from the European Union (EU)’s H2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 643712 to the project Green Bubbles RISE for sustainable diving (Green Bubbles). This paper reflects only the authors’ view. The Research Executive Agency is not responsible for any use that may be made of the information it contains. © 2017. This manuscript version is made available under the CC-BY-NC-ND 4.0 AUTHORS' ACCEPTED MANUSCRIPT Building a baseline for habitat-forming corals by a multi-source approach, including Web Ecological Knowledge - Cristina G Di Camillo, Department of Life and Environmental Sciences, Marche Polytechnic University, CoNISMa, Ancona, Italy, [email protected] - Massimo Ponti, Department of Biological, Geological and Environmental Sciences and Interdepartmental Research Centre for Environmental SciencesUniversity of Bologna, CoNISMa, Ravenna, Italy - Giorgio Bavestrello, Department of Earth, Environment and Life Sciences, University of Genoa, CoNISMa, Genoa, Italy - Maja Krzelj, Department of Marine Studies, University of Split, Split, Croatia - Carlo Cerrano, Department of Life and Environmental Sciences, Marche Polytechnic University, CoNISMa, Ancona, Italy Received: 12 January 2017 Revised: 10 December 2017 Accepted: 14 December 2017 First online: 23 December 2017 Cite as: Di Camillo, C.G., Ponti, M., Bavestrello, G. -

Coelenterata: Anthozoa), with Diagnoses of New Taxa
PROC. BIOL. SOC. WASH. 94(3), 1981, pp. 902-947 KEY TO THE GENERA OF OCTOCORALLIA EXCLUSIVE OF PENNATULACEA (COELENTERATA: ANTHOZOA), WITH DIAGNOSES OF NEW TAXA Frederick M. Bayer Abstract.—A serial key to the genera of Octocorallia exclusive of the Pennatulacea is presented. New taxa introduced are Olindagorgia, new genus for Pseudopterogorgia marcgravii Bayer; Nicaule, new genus for N. crucifera, new species; and Lytreia, new genus for Thesea plana Deich- mann. Ideogorgia is proposed as a replacement ñame for Dendrogorgia Simpson, 1910, not Duchassaing, 1870, and Helicogorgia for Hicksonella Simpson, December 1910, not Nutting, May 1910. A revised classification is provided. Introduction The key presented here was an essential outgrowth of work on a general revisión of the octocoral fauna of the western part of the Atlantic Ocean. The far-reaching zoogeographical affinities of this fauna made it impossible in the course of this study to ignore genera from any part of the world, and it soon became clear that many of them require redefinition according to modern taxonomic standards. Therefore, the type-species of as many genera as possible have been examined, often on the basis of original type material, and a fully illustrated generic revisión is in course of preparation as an essential first stage in the redescription of western Atlantic species. The key prepared to accompany this generic review has now reached a stage that would benefit from a broader and more objective testing under practical conditions than is possible in one laboratory. For this reason, and in order to make the results of this long-term study available, even in provisional form, not only to specialists but also to the growing number of ecologists, biochemists, and physiologists interested in octocorals, the key is now pre- sented in condensed form with minimal illustration. -

Updated Chronology of Mass Mortality Events Hitting Gorgonians in the Western Mediterranean Sea (Modified and Updated from Calvo Et Al
The following supplement accompanies the article Mass mortality hits gorgonian forests at Montecristo Island Eva Turicchia*, Marco Abbiati, Michael Sweet and Massimo Ponti *Corresponding author: [email protected] Diseases of Aquatic Organisms 131: 79–85 (2018) Table S1. Updated chronology of mass mortality events hitting gorgonians in the western Mediterranean Sea (modified and updated from Calvo et al. 2011). Year Locations Scale Depth range Species References (m) 1983 La Ciotat (Ligurian Sea) Local 0 to 20 Eunicella singularis Harmelin 1984 Corallium rubrum 1986 Portofino Promontory (Ligurian Local 0 to 20 Eunicella cavolini Bavestrello & Boero 1988 Sea) 1989 Montecristo Island (Tyrrhenian Local - Paramuricea clavata Guldenschuh in Bavestrello et al. 1994 Sea) 1992 Medes Islands (north-western Local 0 to 14 Paramuricea clavata Coma & Zabala 1992 Mediterranean Sea), Port-Cros 10 to 45 Harmelin & Marinopoulos 1994 National Park 1993 Strait of Messina (Tyrrhenian Local 20 to 39 Paramuricea clavata Mistri & Ceccherelli 1996 Sea), Portofino Promontory Bavestrello et al. 1994 (Ligurian Sea) 1999 Coast of Provence and Ligurian Regional 0 to 45 Paramuricea clavata Cerrano et al. 2000 Sea, Balearic Islands (north- Eunicella singularis Perez et al. 2000 western Mediterranean Sea), Eunicella cavolini Garrabou et al. 2001 Gulf of La Spezia, Port-Cros Eunicella verrucosa Linares et al. 2005 National Park, coast of Calafuria Corallium rubrum Bramanti et al. 2005 (Tyrrhenian Sea) Leptogorgia Coma et al. 2006 sarmentosa Cupido et al. 2008 Crisci et al. 2011 2001 Tavolara Island (Tyrrhenian Sea) Local 10 to 45 Paramuricea Calvisi et al. 2003 clavata Eunicella cavolini 2002 Ischia and Procida Islands Local 15 to 20 Paramuricea clavata Gambi et al. -

Preliminary Report on the Octocorals (Cnidaria: Anthozoa: Octocorallia) from the Ogasawara Islands
国立科博専報,(52), pp. 65–94 , 2018 年 3 月 28 日 Mem. Natl. Mus. Nat. Sci., Tokyo, (52), pp. 65–94, March 28, 2018 Preliminary Report on the Octocorals (Cnidaria: Anthozoa: Octocorallia) from the Ogasawara Islands Yukimitsu Imahara1* and Hiroshi Namikawa2 1Wakayama Laboratory, Biological Institute on Kuroshio, 300–11 Kire, Wakayama, Wakayama 640–0351, Japan *E-mail: [email protected] 2Showa Memorial Institute, National Museum of Nature and Science, 4–1–1 Amakubo, Tsukuba, Ibaraki 305–0005, Japan Abstract. Approximately 400 octocoral specimens were collected from the Ogasawara Islands by SCUBA diving during 2013–2016 and by dredging surveys by the R/V Koyo of the Tokyo Met- ropolitan Ogasawara Fisheries Center in 2014 as part of the project “Biological Properties of Bio- diversity Hotspots in Japan” at the National Museum of Nature and Science. Here we report on 52 lots of these octocoral specimens that have been identified to 42 species thus far. The specimens include seven species of three genera in two families of Stolonifera, 25 species of ten genera in two families of Alcyoniina, one species of Scleraxonia, and nine species of four genera in three families of Pennatulacea. Among them, three species of Stolonifera: Clavularia cf. durum Hick- son, C. cf. margaritiferae Thomson & Henderson and C. cf. repens Thomson & Henderson, and five species of Alcyoniina: Lobophytum variatum Tixier-Durivault, L. cf. mirabile Tixier- Durivault, Lohowia koosi Alderslade, Sarcophyton cf. boletiforme Tixier-Durivault and Sinularia linnei Ofwegen, are new to Japan. In particular, Lohowia koosi is the first discovery since the orig- inal description from the east coast of Australia. -

A Case of Modular Phenotypic Plasticity in the Depth
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Springer - Publisher Connector Calixto-Botía and Sánchez BMC Evolutionary Biology (2017) 17:55 DOI 10.1186/s12862-017-0900-8 RESEARCH ARTICLE Open Access A case of modular phenotypic plasticity in the depth gradient for the gorgonian coral Antillogorgia bipinnata (Cnidaria: Octocorallia) Iván Calixto-Botía1,2* and Juan A. Sánchez2,3 Abstract Background: Phenotypic plasticity, as a phenotypic response induced by the environment, has been proposed as a key factor in the evolutionary history of corals. A significant number of octocoral species show high phenotypic variation, exhibiting a strong overlap in intra- and inter-specific morphologic variation. This is the case of the gorgonian octocoral Antillogorgia bipinnata (Verrill 1864), which shows three polyphyletic morphotypes along a bathymetric gradient. This research tested the phenotypic plasticity of modular traits in A. bipinnata with a reciprocal transplant experiment involving 256 explants from two morphotypes in two locations and at two depths. Vertical and horizontal length and number of new branches were compared 13 weeks following transplant. The data were analysed with a linear mixed-effects model and a graphic approach by reaction norms. Results: At the end of the experiment, 91.8% of explants survived. Lower vertical and horizontal growth rates and lower branch promotion were found for deep environments compared to shallow environments. The overall variation behaved similarly to the performance of native transplants. In particular, promotion of new branches showed variance mainly due to a phenotypic plastic effect. Conclusions: Globally, environmental and genotypic effects explain the variation of the assessed traits. -
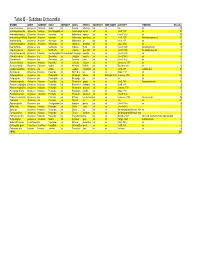
Table B – Subclass Octocorallia
Table B – Subclass Octocorallia BINOMEN ORDER SUBORDER FAMILY SUBFAMILY GENUS SPECIES SUBSPECIES COMN_NAMES AUTHORITY SYNONYMS #Records Acanella arbuscula Alcyonacea Calcaxonia Isididae n/a Acanella arbuscula n/a n/a n/a n/a 59 Acanthogorgia armata Alcyonacea Holaxonia Acanthogorgiidae n/a Acanthogorgia armata n/a n/a Verrill, 1878 n/a 95 Anthomastus agassizii Alcyonacea Alcyoniina Alcyoniidae n/a Anthomastus agassizii n/a n/a (Verrill, 1922) n/a 35 Anthomastus grandiflorus Alcyonacea Alcyoniina Alcyoniidae n/a Anthomastus grandiflorus n/a n/a Verrill, 1878 Anthomastus purpureus 37 Anthomastus sp. Alcyonacea Alcyoniina Alcyoniidae n/a Anthomastus sp. n/a n/a Verrill, 1878 n/a 1 Anthothela grandiflora Alcyonacea Scleraxonia Anthothelidae n/a Anthothela grandiflora n/a n/a (Sars, 1856) n/a 24 Capnella florida Alcyonacea n/a Nephtheidae n/a Capnella florida n/a n/a (Verrill, 1869) Eunephthya florida 44 Capnella glomerata Alcyonacea n/a Nephtheidae n/a Capnella glomerata n/a n/a (Verrill, 1869) Eunephthya glomerata 4 Chrysogorgia agassizii Alcyonacea Holaxonia Acanthogorgiidae Chrysogorgiidae Chrysogorgia agassizii n/a n/a (Verrill, 1883) n/a 2 Clavularia modesta Alcyonacea n/a Clavulariidae n/a Clavularia modesta n/a n/a (Verrill, 1987) n/a 6 Clavularia rudis Alcyonacea n/a Clavulariidae n/a Clavularia rudis n/a n/a (Verrill, 1922) n/a 1 Gersemia fruticosa Alcyonacea Alcyoniina Alcyoniidae n/a Gersemia fruticosa n/a n/a Marenzeller, 1877 n/a 3 Keratoisis flexibilis Alcyonacea Calcaxonia Isididae n/a Keratoisis flexibilis n/a n/a Pourtales, 1868 n/a 1 Lepidisis caryophyllia Alcyonacea n/a Isididae n/a Lepidisis caryophyllia n/a n/a Verrill, 1883 Lepidisis vitrea 13 Muriceides sp. -

Comprehensive Phylogenomic Analyses Resolve Cnidarian Relationships and the Origins of Key Organismal Traits
Comprehensive phylogenomic analyses resolve cnidarian relationships and the origins of key organismal traits Ehsan Kayal1,2, Bastian Bentlage1,3, M. Sabrina Pankey5, Aki H. Ohdera4, Monica Medina4, David C. Plachetzki5*, Allen G. Collins1,6, Joseph F. Ryan7,8* Authors Institutions: 1. Department of Invertebrate Zoology, National Museum of Natural History, Smithsonian Institution 2. UPMC, CNRS, FR2424, ABiMS, Station Biologique, 29680 Roscoff, France 3. Marine Laboratory, university of Guam, UOG Station, Mangilao, GU 96923, USA 4. Department of Biology, Pennsylvania State University, University Park, PA, USA 5. Department of Molecular, Cellular and Biomedical Sciences, University of New Hampshire, Durham, NH, USA 6. National Systematics Laboratory, NOAA Fisheries, National Museum of Natural History, Smithsonian Institution 7. Whitney Laboratory for Marine Bioscience, University of Florida, St Augustine, FL, USA 8. Department of Biology, University of Florida, Gainesville, FL, USA PeerJ Preprints | https://doi.org/10.7287/peerj.preprints.3172v1 | CC BY 4.0 Open Access | rec: 21 Aug 2017, publ: 21 Aug 20171 Abstract Background: The phylogeny of Cnidaria has been a source of debate for decades, during which nearly all-possible relationships among the major lineages have been proposed. The ecological success of Cnidaria is predicated on several fascinating organismal innovations including symbiosis, colonial body plans and elaborate life histories, however, understanding the origins and subsequent diversification of these traits remains difficult due to persistent uncertainty surrounding the evolutionary relationships within Cnidaria. While recent phylogenomic studies have advanced our knowledge of the cnidarian tree of life, no analysis to date has included genome scale data for each major cnidarian lineage. Results: Here we describe a well-supported hypothesis for cnidarian phylogeny based on phylogenomic analyses of new and existing genome scale data that includes representatives of all cnidarian classes. -

Le Coralligène En Méditerranée
Projet pour la préparation d’un Plan d’Action Stratégique pour la Conservation de la Biodiversité dans la Région Méditerranéenne (PAS - BIO) Le coralligène en Méditerranée Définition de la biocénose coralligène en Méditerranée, de ses principaux « constructeurs », de sa richesse et de son rôle en écologie benthique et analyse des principales menaces Projet pour la préparation d’un Plan d’Action Stratégique pour la Conservation de la Biodiversité dans la Région Méditerranéenne (PAS – BIO) Le coralligène en Méditerranée Définition de la biocénose coralligène en Méditerranée, de ses principaux « constructeurs », de sa richesse et de son rôle en écologie benthique et analyse des principales menaces CAR/ASP– Centre d’Activités Régionales pour les Aires Spécialement Protégées 2003 Note : les appellations employées dans ce document et la présentation des données qui y figurent n’impliquent de la part du CAR/ASP et du PNUE aucune prise de position quant au statut juridique des pays, territoires, villes ou zones, ou de leur autorité, ni quant au tracé de leur frontière ou limites. Les avis exprimés dans ce document sont propres à l’auteur et ne représentent pas nécessairement les avis du CAR/ASP ou du PNUE. Ce document a été préparé pour le CAR/ASP par le Dr Enric Ballesteros du Centre d’études Avancées de Blanès (CSIC Accés Cala Sant Francesc, 14. E-17300 Blanes, Girona, Espagne). Sa version française a été légèrement augmentée par Mr Ben Mustapha Karim de l’Institut National des Sciences et Technologies de la Mer (INSTM, Salammbô, Tunisie), avec quelques ajouts relatifs aux données récentes sur le coralligène en Tunisie, afin de donner une idée sur la richesse de cette biocénose en Méditerranée orientale. -

Deep-Sea Life Issue 8, November 2016 Cruise News Going Deep: Deepwater Exploration of the Marianas by the Okeanos Explorer
Deep-Sea Life Issue 8, November 2016 Welcome to the eighth edition of Deep-Sea Life: an informal publication about current affairs in the world of deep-sea biology. Once again we have a wealth of contributions from our fellow colleagues to enjoy concerning their current projects, news, meetings, cruises, new publications and so on. The cruise news section is particularly well-endowed this issue which is wonderful to see, with voyages of exploration from four of our five oceans from the Arctic, spanning north east, west, mid and south Atlantic, the north-west Pacific, and the Indian Ocean. Just imagine when all those data are in OBIS via the new deep-sea node…! (see page 24 for more information on this). The photo of the issue makes me smile. Angelika Brandt from the University of Hamburg, has been at sea once more with her happy-looking team! And no wonder they look so pleased with themselves; they have collected a wonderful array of life from one of the very deepest areas of our ocean in order to figure out more about the distribution of these abyssal organisms, and the factors that may limit their distribution within this region. Read more about the mission and their goals on page 5. I always appreciate feedback regarding any aspect of the publication, so that it may be improved as we go forward. Please circulate to your colleagues and students who may have an interest in life in the deep, and have them contact me if they wish to be placed on the mailing list for this publication. -

Guide to the Identification of Precious and Semi-Precious Corals in Commercial Trade
'l'llA FFIC YvALE ,.._,..---...- guide to the identification of precious and semi-precious corals in commercial trade Ernest W.T. Cooper, Susan J. Torntore, Angela S.M. Leung, Tanya Shadbolt and Carolyn Dawe September 2011 © 2011 World Wildlife Fund and TRAFFIC. All rights reserved. ISBN 978-0-9693730-3-2 Reproduction and distribution for resale by any means photographic or mechanical, including photocopying, recording, taping or information storage and retrieval systems of any parts of this book, illustrations or texts is prohibited without prior written consent from World Wildlife Fund (WWF). Reproduction for CITES enforcement or educational and other non-commercial purposes by CITES Authorities and the CITES Secretariat is authorized without prior written permission, provided the source is fully acknowledged. Any reproduction, in full or in part, of this publication must credit WWF and TRAFFIC North America. The views of the authors expressed in this publication do not necessarily reflect those of the TRAFFIC network, WWF, or the International Union for Conservation of Nature (IUCN). The designation of geographical entities in this publication and the presentation of the material do not imply the expression of any opinion whatsoever on the part of WWF, TRAFFIC, or IUCN concerning the legal status of any country, territory, or area, or of its authorities, or concerning the delimitation of its frontiers or boundaries. The TRAFFIC symbol copyright and Registered Trademark ownership are held by WWF. TRAFFIC is a joint program of WWF and IUCN. Suggested citation: Cooper, E.W.T., Torntore, S.J., Leung, A.S.M, Shadbolt, T. and Dawe, C. -

Deep‐Sea Coral Taxa in the U.S. Gulf of Mexico: Depth and Geographical Distribution
Deep‐Sea Coral Taxa in the U.S. Gulf of Mexico: Depth and Geographical Distribution by Peter J. Etnoyer1 and Stephen D. Cairns2 1. NOAA Center for Coastal Monitoring and Assessment, National Centers for Coastal Ocean Science, Charleston, SC 2. National Museum of Natural History, Smithsonian Institution, Washington, DC This annex to the U.S. Gulf of Mexico chapter in “The State of Deep‐Sea Coral Ecosystems of the United States” provides a list of deep‐sea coral taxa in the Phylum Cnidaria, Classes Anthozoa and Hydrozoa, known to occur in the waters of the Gulf of Mexico (Figure 1). Deep‐sea corals are defined as azooxanthellate, heterotrophic coral species occurring in waters 50 m deep or more. Details are provided on the vertical and geographic extent of each species (Table 1). This list is adapted from species lists presented in ʺBiodiversity of the Gulf of Mexicoʺ (Felder & Camp 2009), which inventoried species found throughout the entire Gulf of Mexico including areas outside U.S. waters. Taxonomic names are generally those currently accepted in the World Register of Marine Species (WoRMS), and are arranged by order, and alphabetically within order by suborder (if applicable), family, genus, and species. Data sources (references) listed are those principally used to establish geographic and depth distribution. Only those species found within the U.S. Gulf of Mexico Exclusive Economic Zone are presented here. Information from recent studies that have expanded the known range of species into the U.S. Gulf of Mexico have been included. The total number of species of deep‐sea corals documented for the U.S. -
![Genetic Divergence and Polyphyly in the Octocoral Genus Swiftia [Cnidaria: Octocorallia], Including a Species Impacted by the DWH Oil Spill](https://docslib.b-cdn.net/cover/9917/genetic-divergence-and-polyphyly-in-the-octocoral-genus-swiftia-cnidaria-octocorallia-including-a-species-impacted-by-the-dwh-oil-spill-739917.webp)
Genetic Divergence and Polyphyly in the Octocoral Genus Swiftia [Cnidaria: Octocorallia], Including a Species Impacted by the DWH Oil Spill
diversity Article Genetic Divergence and Polyphyly in the Octocoral Genus Swiftia [Cnidaria: Octocorallia], Including a Species Impacted by the DWH Oil Spill Janessy Frometa 1,2,* , Peter J. Etnoyer 2, Andrea M. Quattrini 3, Santiago Herrera 4 and Thomas W. Greig 2 1 CSS Dynamac, Inc., 10301 Democracy Lane, Suite 300, Fairfax, VA 22030, USA 2 Hollings Marine Laboratory, NOAA National Centers for Coastal Ocean Sciences, National Ocean Service, National Oceanic and Atmospheric Administration, 331 Fort Johnson Rd, Charleston, SC 29412, USA; [email protected] (P.J.E.); [email protected] (T.W.G.) 3 Department of Invertebrate Zoology, National Museum of Natural History, Smithsonian Institution, 10th and Constitution Ave NW, Washington, DC 20560, USA; [email protected] 4 Department of Biological Sciences, Lehigh University, 111 Research Dr, Bethlehem, PA 18015, USA; [email protected] * Correspondence: [email protected] Abstract: Mesophotic coral ecosystems (MCEs) are recognized around the world as diverse and ecologically important habitats. In the northern Gulf of Mexico (GoMx), MCEs are rocky reefs with abundant black corals and octocorals, including the species Swiftia exserta. Surveys following the Deepwater Horizon (DWH) oil spill in 2010 revealed significant injury to these and other species, the restoration of which requires an in-depth understanding of the biology, ecology, and genetic diversity of each species. To support a larger population connectivity study of impacted octocorals in the Citation: Frometa, J.; Etnoyer, P.J.; GoMx, this study combined sequences of mtMutS and nuclear 28S rDNA to confirm the identity Quattrini, A.M.; Herrera, S.; Greig, Swiftia T.W.