Supplemental Figures.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Neuromodulation of Whisking Related Neural Activity in Superior Colliculus
The Journal of Neuroscience, May 28, 2014 • 34(22):7683–7695 • 7683 Systems/Circuits Neuromodulation of Whisking Related Neural Activity in Superior Colliculus Tatiana Bezdudnaya and Manuel A. Castro-Alamancos Department of Neurobiology and Anatomy, Drexel University College of Medicine, Philadelphia, Pennsylvania 19129 The superior colliculus is part of a broader neural network that can decode whisker movements in air and on objects, which is a strategy used by behaving rats to sense the environment. The intermediate layers of the superior colliculus receive whisker-related excitatory afferents from the trigeminal complex and barrel cortex, inhibitory afferents from extrinsic and intrinsic sources, and neuromodulatory afferents from cholinergic and monoaminergic nuclei. However, it is not well known how these inputs regulate whisker-related activity in the superior colliculus. We found that barrel cortex afferents drive the superior colliculus during the middle portion of the rising phase of the whisker movement protraction elicited by artificial (fictive) whisking in anesthetized rats. In addition, both spontaneous and whisker-related neural activities in the superior colliculus are under strong inhibitory and neuromodulator control. Cholinergic stimu- lation activates the superior colliculus by increasing spontaneous firing and, in some cells, whisker-evoked responses. Monoaminergic stimulation has the opposite effects. The actions of neuromodulator and inhibitory afferents may be the basis of the different firing rates and sensory responsiveness observed in the superior colliculus of behaving animals during distinct behavioral states. Key words: acetylcholine; neuromodulation; norepinephrine; superior colliculus; whisker Introduction ment and active touch (i.e., whisking on objects; Bezdudnaya and The superior colliculus is a well known hub for sensorimotor Castro-Alamancos, 2011). -

The Superior Colliculus–Pretectum Mediates the Direct Effects of Light on Sleep
Proc. Natl. Acad. Sci. USA Vol. 95, pp. 8957–8962, July 1998 Neurobiology The superior colliculus–pretectum mediates the direct effects of light on sleep ANN M. MILLER*, WILLIAM H. OBERMEYER†,MARY BEHAN‡, AND RUTH M. BENCA†§ *Neuroscience Training Program and †Department of Psychiatry, University of Wisconsin–Madison, 6001 Research Park Boulevard, Madison, WI 53719; and ‡Department of Comparative Biosciences, University of Wisconsin–Madison, Room 3466, Veterinary Medicine Building, 2015 Linden Drive West, Madison, WI 53706 Communicated by James M. Sprague, The University of Pennsylvania School of Medicine, Philadelphia, PA, May 27, 1998 (received for review August 26, 1997) ABSTRACT Light and dark have immediate effects on greater REM sleep expression occurring in light rather than sleep and wakefulness in mammals, but the neural mecha- dark periods (8, 9). nisms underlying these effects are poorly understood. Lesions Another behavioral response of nocturnal rodents to of the visual cortex or the superior colliculus–pretectal area changes in lighting conditions consists of increased amounts of were performed in albino rats to determine retinorecipient non-REM (NREM) sleep and total sleep after lights-on and areas that mediate the effects of light on behavior, including increased wakefulness following lights-off (4). None of the rapid eye movement sleep triggering by lights-off and redis- light-induced behaviors (i.e., REM sleep, NREM sleep, or tribution of non-rapid eye movement sleep in short light–dark waking responses to lighting changes) appears to be under cycles. Acute responses to changes in light conditions were primary circadian control: the behaviors are not eliminated by virtually eliminated by superior colliculus-pretectal area le- destruction of the suprachiasmatic nucleus (24) and can be sions but not by visual cortex lesions. -

The Superior and Inferior Colliculi of the Mole (Scalopus Aquaticus Machxinus)
THE SUPERIOR AND INFERIOR COLLICULI OF THE MOLE (SCALOPUS AQUATICUS MACHXINUS) THOMAS N. JOHNSON' Laboratory of Comparative Neurology, Departmmt of Amtomy, Un&versity of hfiehigan, Ann Arbor INTRODUCTION This investigation is a study of the afferent and efferent connections of the tectum of the midbrain in the mole (Scalo- pus aquaticus machrinus). An attempt is made to correlate these findings with the known habits of the animal. A subterranean animal of the middle western portion of the United States, Scalopus aquaticus machrinus is the largest of the genus Scalopus and its habits have been more thor- oughly studied than those of others of this genus according to Jackson ('15) and Hamilton ('43). This animal prefers a well-drained, loose soil. It usually frequents open fields and pastures but also is found in thin woods and meadows. Following a rain, new superficial burrows just below the surface of the ground are pushed in all directions to facili- tate the capture of worms and other soil life. Ten inches or more below the surface the regular permanent highway is constructed; the mole retreats here during long periods of dry weather or when frost is in the ground. The principal food is earthworms although, under some circumstances, larvae and adult insects are the more usual fare. It has been demonstrated conclusively that, under normal conditions, moles will eat vegetable matter. It seems not improbable that they may take considerable quantities of it at times. A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the University of Michigan. -

VISUAL RESPONSE PROPERTIES in the TECTORECIPIENT ZONE of the CAT’S LATERAL POSTERIOR-PULVINAR COMPLEX: a COMPARISON with the SUPERIOR Colliculusl
0270-6474/82/0312-2587$02.00/O The Journal of Neuroscience Copyright 0 Society for Neuroscience Vol. 3, No. 12, pp. 2587-2596 Printed in U.S.A. December 1983 VISUAL RESPONSE PROPERTIES IN THE TECTORECIPIENT ZONE OF THE CAT’S LATERAL POSTERIOR-PULVINAR COMPLEX: A COMPARISON WITH THE SUPERIOR coLLIcuLusl LEO M. CHALUPA,’ ROBERT W. WILLIAMS,3 AND MICHAEL J. HUGHES Department of Psychology and The Physiology Graduate Group, University of California, Davis, California 95616 Received April 4, 1983; Revised June 16,1983; Accepted June 28, 1983 Abstract The medial portion of the cat’s lateral posterior-pulvinar complex (LPm) receives a prominent ascending projection from the superficial layers of the superior colliculus. This region of the thalamus has been suggested to serve as a relay by which visual information from the midbrain could be conveyed to extrastriate cortex. In order to determine how the functional organization within the LPm compares with that of the superior colliculus, visual response properties of LPm and superior collicular neurons were examined under identical experimental conditions. The majority of neurons in the LPm, as in the superior colliculus, respond vigorously to moving stimuli, and a substantial proportion of these cells also exhibit a preference for movements in a particular direction. Further- more, most cells in the LPm, in common with those of the tectum, respond only in a phasic manner to flashed stimuli, have homogeneous receptive field organization, and show response suppression to stimuli larger than the activating region of the receptive field. As in the colliculus, the ipsilateral visual field is represented in the LPm. -
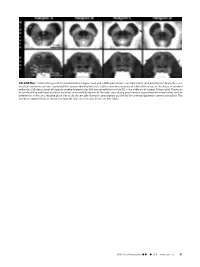
ON-LINE FIG 1. Selected Images of the Caudal Midbrain (Upper Row
ON-LINE FIG 1. Selected images of the caudal midbrain (upper row) and middle pons (lower row) from 4 of 13 total postmortem brains illustrate excellent anatomic contrast reproducibility across individual datasets. Subtle variations are present. Note differences in the shape of cerebral peduncles (24), decussation of superior cerebellar peduncles (25), and spinothalamic tract (12) in the midbrain of subject D (top right). These can be attributed to individual anatomic variation, some mild distortion of the brain stem during procurement at postmortem examination, and/or differences in the axial imaging plane not easily discernable during its prescription parallel to the anterior/posterior commissure plane. The numbers in parentheses in the on-line legends refer to structures in the On-line Table. AJNR Am J Neuroradiol ●:●●2019 www.ajnr.org E1 ON-LINE FIG 3. Demonstration of the dentatorubrothalamic tract within the superior cerebellar peduncle (asterisk) and rostral brain stem. A, Axial caudal midbrain image angled 10° anterosuperior to posteroinferior relative to the ACPC plane demonstrates the tract traveling the midbrain to reach the decussation (25). B, Coronal oblique image that is perpendicular to the long axis of the hippocam- pus (structure not shown) at the level of the ventral superior cerebel- lar decussation shows a component of the dentatorubrothalamic tract arising from the cerebellar dentate nucleus (63), ascending via the superior cerebellar peduncle to the decussation (25), and then enveloping the contralateral red nucleus (3). C, Parasagittal image shows the relatively long anteroposterior dimension of this tract, which becomes less compact and distinct as it ascends toward the thalamus. ON-LINE FIG 2. -

Distinct Higher-Order Thalamic Circuits Channel Parallel Streams of Visual Information in Mice
bioRxiv preprint doi: https://doi.org/10.1101/395244; this version posted August 18, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Distinct higher-order thalamic circuits channel parallel streams of visual information in mice Corbett Bennett1*, Samuel D. Gale1*#, Marina E. Garrett1,2, Melissa L. Newton2, Edward M. Callaway2, Gabe J. Murphy1, Shawn R. Olsen1 1. Allen Institute for Brain Science 2. Salk Institute for Biological Studies * contributed equally # corresponding author ([email protected]) MLN’s present affiliation is the University of California at Berkeley Abstract Higher-order thalamic nuclei, such as the visual pulvinar, play essential roles in shaping cortical dynamics and connecting functionally-related cortical and subcortical brain regions. A coherent framework describing pulvinar function remains elusive due to its anatomical complexity, involvement in diverse cognitive processes, and the limited experimental tools available in many species. We combined large-scale anatomical circuit mapping with high-density electrophysiological recordings to dissect a homolog of pulvinar in mice, the lateral posterior nucleus (LP). We define three LP subregions based on correspondence between connectivity and functional properties. These subregions form parallel corticothalamic loops and contain separate representations of visual space. Silencing visual cortex or the superior colliculus revealed that these input sources drive activity and shape visual tuning in separate LP subregions. By specifying the information carried by distinct circuits through LP and identifying their downstream targets, our data provide a roadmap for understanding pulvinar function in visual processing and behavior. Introduction Higher-order thalamus plays a critical role in cortical function, both as a route by which cortical areas communicate and a relay of subcortical input to cortex. -

Architectonic Characteristics of the Visual Thalamus and Superior Colliculus in Titi Monkeys
Received: 23 December 2017 | Revised: 21 March 2018 | Accepted: 22 March 2018 DOI: 10.1002/cne.24445 RESEARCH ARTICLE Architectonic characteristics of the visual thalamus and superior colliculus in titi monkeys Mary K L Baldwin | Leah Krubitzer Center for Neuroscience, University of California, 1544 Newton Court, Davis, Abstract California Titi monkeys are arboreal, diurnal New World monkeys whose ancestors were the first surviving branch of the New World radiation. In the current study, we use cytoarchitectonic and immunohis- Correspondence tochemical characteristics to compare titi monkey subcortical structures associated with visual Mary K L Baldwin, University of California, Davis, Center for Neuroscience, 1544 processing with those of other well-studied primates. Our goal was to appreciate features that are Newton Court, Davis, CA 95618. similar across all New World monkeys, and primates in general, versus those features that are Email: [email protected] unique to titi monkeys and other primate taxa. We examined tissue stained for Nissl substance, cytochrome oxidase (CO), acetylcholinesterase (AChE), calbindin (Cb), parvalbumin (Pv), and vesicu- lar glutamate transporter 2 (VGLUT2) to characterize the superior colliculus, lateral geniculate nucleus, and visual pulvinar. This is the first study to characterize VGLUT2 in multiple subcortical structures of any New World monkey. Our results from tissue processed for VGLUT2, in combina- tion with other histological stains, revealed distinct features of subcortical structures that are similar to other primates, but also some features that are slightly modified compared to other New World monkeys and other primates. These included subdivisions of the inferior pulvinar, sublamina within the stratum griseum superficiale (SGS) of the superior colliculus, and specific koniocellular layers within the lateral geniculate nucleus. -

Brain Anatomy
BRAIN ANATOMY Adapted from Human Anatomy & Physiology by Marieb and Hoehn (9th ed.) The anatomy of the brain is often discussed in terms of either the embryonic scheme or the medical scheme. The embryonic scheme focuses on developmental pathways and names regions based on embryonic origins. The medical scheme focuses on the layout of the adult brain and names regions based on location and functionality. For this laboratory, we will consider the brain in terms of the medical scheme (Figure 1): Figure 1: General anatomy of the human brain Marieb & Hoehn (Human Anatomy and Physiology, 9th ed.) – Figure 12.2 CEREBRUM: Divided into two hemispheres, the cerebrum is the largest region of the human brain – the two hemispheres together account for ~ 85% of total brain mass. The cerebrum forms the superior part of the brain, covering and obscuring the diencephalon and brain stem similar to the way a mushroom cap covers the top of its stalk. Elevated ridges of tissue, called gyri (singular: gyrus), separated by shallow groves called sulci (singular: sulcus) mark nearly the entire surface of the cerebral hemispheres. Deeper groves, called fissures, separate large regions of the brain. Much of the cerebrum is involved in the processing of somatic sensory and motor information as well as all conscious thoughts and intellectual functions. The outer cortex of the cerebrum is composed of gray matter – billions of neuron cell bodies and unmyelinated axons arranged in six discrete layers. Although only 2 – 4 mm thick, this region accounts for ~ 40% of total brain mass. The inner region is composed of white matter – tracts of myelinated axons. -

Movements Resembling Orientation Or Avoidance Elicited by Electrical Stimulation of the Superior Colliculus in Rats
The Journal of Neuroscience March 1986, 6(3): 723-733 Movements Resembling Orientation or Avoidance Elicited by Electrical Stimulation of the Superior Colliculus in Rats Niar Sahibzada, Paul Dean, and Peter Redgrave Department of Psychology, University of Sheffield, Sheffield SlO 2TN, England Some studies have reported that stimulation of the superior col- out on cats and monkeys, in rats too collicular stimulation can liculus in rats produces orienting responses, as it does in a num- induce movements of the eyes, head, and body that resemble ber of species. However, other studies have reported movements orientation and approach (lmperato and Di Chiara, 198 1; Kil- resembling avoidance and escape, which are not characteristic patrick et al., 1982; McHaffie and Stein, 1982; Weldon et al., of collicular stimulation in other mammals. This apparent dis- 1983). crepancy was investigated by systematically recording the ef- However, these are not the only movements that have been fects on head and body movements of electrical stimulation at reported to follow collicular stimulation in rats. In a number of a large number of sites throughout the superior colliculus (SC) studies, electrical stimulation of the rat SC has produced escape and surrounding structures. or other responses characteristic of aversive stimulation (Olds It was found that the nature of the movements observed de- and Olds, 1962, 1963; Olds and Peretz, 1960; Schmitt et al., pended on the location of the stimulating electrode. Contralat- 1974; Stein, 1965; Valenstein, 1965; Waldbillig, 1975). Because era1 head and body movements resembling orienting and ap- electrical stimulation affects axons as well as cell bodies and proach were obtained from sites in the intermediate and deep dendrites, the substrate of these escape or avoidance responses layers in rostra1 colliculus, the intermediate white layer and could have been either fibers passing through the SC or the immediately surrounding tissue in central colliculus, and in all collaterals of fibers that terminate elsewhere in the brain. -

The Role of the Superior Colliculus in Visually Guided Locomotion and Visual Orienting in the Hamster
Physiological Psychology 1980, Vol. 8 (1),20·28 The role of the superior colliculus in visually guided locomotion and visual orienting in the hamster ELIZABETH MORT, SARA CAIRNS, HELEN HERSCH, and BARBARA FINLAY Cornell University, Ithaca, New York 14853 Visually guided locomotion toward goal doors designated by various cues was assessed in normal hamsters, hamsters with undercuts of the superior colliculus, and hamsters with neonatal lesions of the superior colliculus. Scanning of visual arrays, orientation to novel stimuli, and orientation to sunflower seeds were assessed in the same animals. Collicular lesions produced no deficit in accuracy of choice of goal door, type of route taken, or speed of locomotion. By contrast, the same hamsters showed major deficits in spontaneous scan· ning and orientation to stimuli in the visual periphery. Hamsters with neonatal and adult collicular lesions performed identically on every task except orientation to sunflower seeds presented at various locations in the visual field; adult operates were somewhat more vari· able in their performance. The implications of these results for the concept of encephalization of function and the phylogeny of the optic tectum are discussed. In every vertebrate that has been studied, the mid 1969, 1970) has shown a deficit both in visu1ll orienting, brain tectum plays some role in visuomotor behavior as measured by head turns to sunflower seeds, and in (Ingle & Sprague, 1975). In highly visual animals visually guided locomotion consequent to a collicular with marked retinal specializations for pattern vision, undercut, and has characterized these symtoms as the superior colliculus has been implicated in acqui indicative of a general disability in spatial localization, sition of visual targets, particularly peripheral ones knowing where things are. -

Focused Ultrasound Neuromodulation of Cortical and Subcortical Brain Structures Using 1.9 Mhz Hermes A
Focused ultrasound neuromodulation of cortical and subcortical brain structures using 1.9 MHz Hermes A. S. Kamimura, Shutao Wang, Hong Chen, Qi Wang, Christian Aurup, Camilo Acosta, Antonio A. O. Carneiro, and Elisa E. Konofagou Citation: Medical Physics 43, 5730 (2016); doi: 10.1118/1.4963208 View online: http://dx.doi.org/10.1118/1.4963208 View Table of Contents: http://scitation.aip.org/content/aapm/journal/medphys/43/10?ver=pdfcov Published by the American Association of Physicists in Medicine Articles you may be interested in Targeting accuracy and closing timeline of the microbubble-enhanced focused ultrasound blood-brain barrier opening in non-human primates AIP Conf. Proc. 1503, 35 (2012); 10.1063/1.4769913 Neuronavigation-guided focused ultrasound-induced blood-brain barrier opening: A preliminary study in swine AIP Conf. Proc. 1503, 29 (2012); 10.1063/1.4769912 Mechanical bioeffects of pulsed high intensity focused ultrasound on a simple neural model Med. Phys. 39, 4274 (2012); 10.1118/1.4729712 An anthropomorphic polyvinyl alcohol brain phantom based on Colin27 for use in multimodal imaging Med. Phys. 39, 554 (2012); 10.1118/1.3673069 The mechanism of interaction between focused ultrasound and microbubbles in blood-brain barrier opening in mice J. Acoust. Soc. Am. 130, 3059 (2011); 10.1121/1.3646905 Focused ultrasound neuromodulation of cortical and subcortical brain structures using 1.9 MHz Hermes A. S. Kamimuraa) Department of Biomedical Engineering, Columbia University, New York, New York 10027 and Department of Physics, FFCLRP, University of São Paulo, Ribeirão Preto, SP 14040-901, Brazil Shutao Wang, Hong Chen, Qi Wang, Christian Aurup, and Camilo Acosta Department of Biomedical Engineering, Columbia University, New York, New York 10027 Antonio A. -

Brainstem Inputs to the Ferret Medial Geniculate Nucleus and the Effect of Early Deafferentation on Novel Retinal Projections to the Auditory Thalamus
THE JOURNAL OF COMPARATIVE NEUROLOGY 400:417–439 (1998) Brainstem Inputs to the Ferret Medial Geniculate Nucleus and the Effect of Early Deafferentation on Novel Retinal Projections to the Auditory Thalamus ALESSANDRA ANGELUCCI, FRANCISCO CLASCA´ , AND MRIGANKA SUR* Department of Brain and Cognitive Sciences, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139 ABSTRACT Following specific neonatal brain lesions in rodents and ferrets, retinal axons have been induced to innervate the medial geniculate nucleus (MGN). Previous studies have suggested that reduction of normal retinal targets along with deafferentation of the MGN are two concurrent factors required for the induction of novel retino-MGN projections. We have examined, in ferrets, the relative influence of these two factors on the extent of the novel retinal projection. We first characterized the inputs to the normal MGN, and the most effective combination of neonatal lesions to deafferent this nucleus, by injecting retrograde tracers into the MGN of normal and neonatally operated adult ferrets, respectively. In a second group of experiments, newborn ferrets received different combinations of lesions of normal retinal targets and MGN afferents. The resulting extent of retino-MGN projections was estimated for each case at adulthood, by using intraocular injections of anterograde tracers. We found that the extent of retino-MGN projections correlates well with the extent of MGN deafferentation, but not with extent of removal of normal retinal targets. Indeed, the presence of at least some normal retinal targets seems necessary for the formation of retino-MGN connections. The diameters of retino-MGN axons suggest that more than one type of retinal ganglion cells innervate the MGN under a lesion paradigm that spares the visual cortex and lateral geniculate nucleus.