ADAM23 Is Downregulated in Side Population and Suppresses Lung Metastasis of Lung Carcinoma Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Peking University-Juntendo University Joint Symposium on Cancer Research and Treatment ADAM28 (A Disintegrin and Metalloproteinase 28) in Cancer Cell Proliferation and Progression
Whatʼs New from Juntendo University, Tokyo Juntendo Medical Journal 2017. 63(5), 322-325 Peking University - Juntendo University Joint Symposium on Cancer Research and Treatment ADAM28 (a Disintegrin and Metalloproteinase 28) in Cancer Cell Proliferation and Progression YASUNORI OKADA* *Department of Pathophysiology for Locomotive and Neoplastic Diseases, Juntendo University Graduate School of Medicine, Tokyo, Japan A disintegrinandmetalloproteinase 28 (ADAM28) is overexpressedpredominantlyby carcinoma cells in more than 70% of the non-small cell lung carcinomas, showing positive correlations with carcinoma cell proliferation and metastasis. ADAM28 cleaves insulin-like growth factor binding protein-3 (IGFBP-3) in the IGF-I/IGFBP-3 complex, leading to stimulation of cell proliferation by intact IGF-I released from the complex. ADAM28 also degrades von Willebrand factor (VWF), which induces apoptosis in human carcinoma cell lines with negligible ADAM28 expression, andthe VWF digestionby ADAM28-expressing carcinoma cells facilitates them to escape from VWF-induced apoptosis, resulting in promotion of metastasis. We have developed human antibodies against ADAM28 andshown that one of them significantly inhibits tumor growth andmetastasis using lung adenocarcinoma cells. Our data suggest that ADAM28 may be a new molecular target for therapy of the patients with ADAM28-expressing non-small cell lung carcinoma. Key words: a disintegrin and metalloproteinase 28 (ADAM28), cell proliferation, invasion, metastasis, human antibody inhibitor Introduction human cancers 2). However, development of the synthetic inhibitors of MMPs andtheir application Cancer cell proliferation andprogression are for treatment of the cancer patients failed 3). modulated by proteolytic cleavage of tissue micro- On the other hand, members of the ADAM (a environmental factors such as extracellular matrix disintegrin and metalloproteinase) gene family, (ECM), growth factors andcytokines, receptors another family belonging to the metzincin gene andcell adhesionmolecules. -

Human ADAM12 Quantikine ELISA
Quantikine® ELISA Human ADAM12 Immunoassay Catalog Number DAD120 For the quantitative determination of A Disintegrin And Metalloproteinase domain- containing protein 12 (ADAM12) concentrations in cell culture supernates, serum, plasma, and urine. This package insert must be read in its entirety before using this product. For research use only. Not for use in diagnostic procedures. TABLE OF CONTENTS SECTION PAGE INTRODUCTION .....................................................................................................................................................................1 PRINCIPLE OF THE ASSAY ...................................................................................................................................................2 LIMITATIONS OF THE PROCEDURE .................................................................................................................................2 TECHNICAL HINTS .................................................................................................................................................................2 MATERIALS PROVIDED & STORAGE CONDITIONS ...................................................................................................3 OTHER SUPPLIES REQUIRED .............................................................................................................................................3 PRECAUTIONS .........................................................................................................................................................................4 -

Conservation and Divergence of ADAM Family Proteins in the Xenopus Genome
Wei et al. BMC Evolutionary Biology 2010, 10:211 http://www.biomedcentral.com/1471-2148/10/211 RESEARCH ARTICLE Open Access ConservationResearch article and divergence of ADAM family proteins in the Xenopus genome Shuo Wei*1, Charles A Whittaker2, Guofeng Xu1, Lance C Bridges1,3, Anoop Shah1, Judith M White1 and Douglas W DeSimone1 Abstract Background: Members of the disintegrin metalloproteinase (ADAM) family play important roles in cellular and developmental processes through their functions as proteases and/or binding partners for other proteins. The amphibian Xenopus has long been used as a model for early vertebrate development, but genome-wide analyses for large gene families were not possible until the recent completion of the X. tropicalis genome sequence and the availability of large scale expression sequence tag (EST) databases. In this study we carried out a systematic analysis of the X. tropicalis genome and uncovered several interesting features of ADAM genes in this species. Results: Based on the X. tropicalis genome sequence and EST databases, we identified Xenopus orthologues of mammalian ADAMs and obtained full-length cDNA clones for these genes. The deduced protein sequences, synteny and exon-intron boundaries are conserved between most human and X. tropicalis orthologues. The alternative splicing patterns of certain Xenopus ADAM genes, such as adams 22 and 28, are similar to those of their mammalian orthologues. However, we were unable to identify an orthologue for ADAM7 or 8. The Xenopus orthologue of ADAM15, an active metalloproteinase in mammals, does not contain the conserved zinc-binding motif and is hence considered proteolytically inactive. We also found evidence for gain of ADAM genes in Xenopus as compared to other species. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Differential Gene Expression and Functional Analysis Implicate Novel Mechanisms in Enteric Nervous System Precursor Migration and Neuritogenesis
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Developmental Biology 298 (2006) 259–271 www.elsevier.com/locate/ydbio Differential gene expression and functional analysis implicate novel mechanisms in enteric nervous system precursor migration and neuritogenesis Bhupinder P.S. Vohra a, Keiji Tsuji b,c, Mayumi Nagashimada b,d, Toshihiro Uesaka b, ⁎ ⁎ Daniel Wind a, Ming Fu a, Jennifer Armon a, Hideki Enomoto b, , Robert O. Heuckeroth a, a Department of Pediatrics and Department of Molecular Biology and Pharmacology, Washington University School of Medicine, 660 S. Euclid Avenue, Box 8208, St. Louis, MO 63110, USA b Laboratory for Neuronal Differentiation and Regeneration, RIKEN Center for Developmental Biology, 2-2-3 Minatojima-minamimachi, Chuo-ku, Kobe, Hyogo 650-0047, Japan c Department of Pediatrics, Kyoto Prefectural University of Medicine, Graduate School of Medical Science, Kamigyo-ku, Kyoto 602-8566, Japan d Osaka University Graduate School of Medicine, 2-2 Yamadaoka, Suita, Osaka 565-0871, Japan Received for publication 17 April 2006; revised 17 May 2006; accepted 22 June 2006 Available online 27 June 2006 Abstract Enteric nervous system (ENS) development requires complex interactions between migrating neural-crest-derived cells and the intestinal microenvironment. Although some molecules influencing ENS development are known, many aspects remain poorly understood. To identify additional molecules critical for ENS development, we used DNA microarray, quantitative real-time PCR and in situ hybridization to compare gene expression in E14 and P0 aganglionic or wild type mouse intestine. Eighty-three genes were identified with at least two-fold higher expression in wild type than aganglionic bowel. -

Comparative Transcriptome Analyses Reveal Genes Associated with SARS-Cov-2 Infection of Human Lung Epithelial Cells
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.24.169268; this version posted June 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Comparative transcriptome analyses reveal genes associated with SARS-CoV-2 infection of human lung epithelial cells Darshan S. Chandrashekar1, *, Upender Manne1,2,#, Sooryanarayana Varambally1,2,3,#* 1Department of Pathology, University of Alabama at Birmingham, Birmingham, AL 2Comprehensive Cancer Center, University of Alabama at Birmingham, Birmingham, AL 3Institute of Informatics, University of Alabama at Birmingham, Birmingham, AL # Share Senior Authorship (UM Email: [email protected]) *Correspondence to: Sooryanarayana Varambally, Ph.D., Molecular and Cellular Pathology, Department of Pathology, Wallace Tumor Institute, 4th floor, 20B, University of Alabama at Birmingham, Birmingham, AL 35233, USA Phone: (205) 996-1654 Email: [email protected] And Darshan S. Chandrashekar Ph.D., Department of Pathology, University of Alabama at Birmingham, Birmingham, AL Email: [email protected] Running Title: SARS-CoV-2 gene signature in infected lung epithelial cells Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed. Page | 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.24.169268; this version posted June 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract: Understanding the molecular mechanism of SARS-CoV-2 infection (the cause of COVID-19) is a scientific priority for 2020. Various research groups are working toward development of vaccines and drugs, and many have published genomic and transcriptomic data related to this viral infection. -
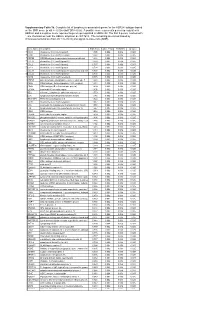
* Supplementary Table 3B. Complete List of Lymphocytic-Associated Genes
Supplementary Table 3b. Complete list of lymphocytic-associated genes for the HER2+I subtype based on the SNR score (p-val <= 0.005 and FDR<=0.05). A positive score represents genes up-regulated in HER2+I and a negative score represents genes up-regulated in HER2+NI. The first 9 genes, marked with *, are chemokines near the HER2+ amplicon at chr17q12. The remaining are sorted based by chromosomal location (from chr 1 to chr X) and signal-to-noise ratio (SNR). Gene Name Description SNR Score Feature P value FDR(BH) Q Value * CCL5 chemokine (C-C motif) ligand 5 1.395 0.002 0.036 0.025 * CCR7 chemokine (C-C motif) receptor 7 1.362 0.002 0.036 0.025 * CD79B CD79B antigen (immunoglobulin-associated beta) 1.248 0.002 0.036 0.025 * CCL13 chemokine (C-C motif) ligand 13 1.003 0.002 0.036 0.025 * CCL2 chemokine (C-C motif) ligand 2 0.737 0.004 0.056 0.037 * CCL8 chemokine (C-C motif) ligand 8 0.724 0.002 0.036 0.025 * CCL18 chemokine (C-C motif) ligand 18 (pulmonary and activ 0.703 0.002 0.036 0.025 * CCL23 chemokine (C-C motif) ligand 23 0.701 0.002 0.036 0.025 * CCR6 chemokine (C-C motif) receptor 6 0.715 0.002 0.036 0.025 PTPN7 protein tyrosine phosphatase, non-receptor type 7 1.861 0.002 0.036 0.025 CD3Z CD3Z antigen, zeta polypeptide (TiT3 complex) 1.811 0.002 0.036 0.025 CD48 CD48 antigen (B-cell membrane protein) 1.809 0.002 0.036 0.025 IL10RA interleukin 10 receptor, alpha 1.806 0.002 0.036 0.025 SELL selectin L (lymphocyte adhesion molecule 1) 1.773 0.002 0.036 0.025 LCK lymphocyte-specific protein tyrosine kinase 1.745 0.002 0.036 0.025 -

Microenvironment-Derived ADAM28 Prevents Cancer Dissemination
www.oncotarget.com Oncotarget, 2018, Vol. 9, (No. 98), pp: 37185-37199 Research Paper Microenvironment-derived ADAM28 prevents cancer dissemination Catherine Gérard1, Céline Hubeau1, Oriane Carnet1, Marine Bellefroid1, Nor Eddine Sounni1, Silvia Blacher1, Guillaume Bendavid1,2, Markus Moser3, Reinhard Fässler3, Agnès Noel1, Didier Cataldo1,4,* and Natacha Rocks1,* 1Laboratory of Tumor and Development Biology, GIGA-Cancer and GIGA-I3, GIGA-Research, University of Liege, Liege, Belgium 2ENT Department, University Hospital of Liege, Liege, Belgium 3Max-Planck-Institute of Biochemistry, Department of Molecular Medicine, Martinsried, Germany 4Department of Respiratory Diseases, CHU Liege and University of Liege, Liege, Belgium *These authors have contributed equally to this work Correspondence to: Didier Cataldo, email: [email protected] Keywords: ADAM28; lung; metastasis; CD8+; T lymphocytes Received: July 27, 2018 Accepted: November 26, 2018 Published: December 14, 2018 Copyright: Gérard et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Previous studies have linked cancer cell-associated ADAM28 expression with tumor progression and metastatic dissemination. However, the role of host-derived ADAM28 in cancer dissemination processes remains unclear. Genetically engineered-mice fully deficient for ADAM28 unexpectedly display increased lung colonization by pulmonary, melanoma or breast tumor cells. In experimental tumor cell dissemination models, host ADAM28 deficiency is further associated with a decreased lung infiltration by CD8+ T lymphocytes. Notably, naive ADAM28-deficient mice already display a drastic reduction of CD8+ T cells in spleen which is further observed in lungs. -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

Molecular Signatures Differentiate Immune States in Type 1 Diabetes Families
Page 1 of 65 Diabetes Molecular signatures differentiate immune states in Type 1 diabetes families Yi-Guang Chen1, Susanne M. Cabrera1, Shuang Jia1, Mary L. Kaldunski1, Joanna Kramer1, Sami Cheong2, Rhonda Geoffrey1, Mark F. Roethle1, Jeffrey E. Woodliff3, Carla J. Greenbaum4, Xujing Wang5, and Martin J. Hessner1 1The Max McGee National Research Center for Juvenile Diabetes, Children's Research Institute of Children's Hospital of Wisconsin, and Department of Pediatrics at the Medical College of Wisconsin Milwaukee, WI 53226, USA. 2The Department of Mathematical Sciences, University of Wisconsin-Milwaukee, Milwaukee, WI 53211, USA. 3Flow Cytometry & Cell Separation Facility, Bindley Bioscience Center, Purdue University, West Lafayette, IN 47907, USA. 4Diabetes Research Program, Benaroya Research Institute, Seattle, WA, 98101, USA. 5Systems Biology Center, the National Heart, Lung, and Blood Institute, the National Institutes of Health, Bethesda, MD 20824, USA. Corresponding author: Martin J. Hessner, Ph.D., The Department of Pediatrics, The Medical College of Wisconsin, Milwaukee, WI 53226, USA Tel: 011-1-414-955-4496; Fax: 011-1-414-955-6663; E-mail: [email protected]. Running title: Innate Inflammation in T1D Families Word count: 3999 Number of Tables: 1 Number of Figures: 7 1 For Peer Review Only Diabetes Publish Ahead of Print, published online April 23, 2014 Diabetes Page 2 of 65 ABSTRACT Mechanisms associated with Type 1 diabetes (T1D) development remain incompletely defined. Employing a sensitive array-based bioassay where patient plasma is used to induce transcriptional responses in healthy leukocytes, we previously reported disease-specific, partially IL-1 dependent, signatures associated with pre and recent onset (RO) T1D relative to unrelated healthy controls (uHC). -

Mutation Analysis of Candidate Genes Within the 2Q33.3 Linkage Area for Familial Early-Onset Generalised Osteoarthritis
European Journal of Human Genetics (2007) 15, 791–799 & 2007 Nature Publishing Group All rights reserved 1018-4813/07 $30.00 www.nature.com/ejhg ARTICLE Mutation analysis of candidate genes within the 2q33.3 linkage area for familial early-onset generalised osteoarthritis Josine L Min1, Ingrid Meulenbelt*,1, Margreet Kloppenburg2, Cornelia M van Duijn3 and P Eline Slagboom1 1Molecular Epidemiology, Leiden University Medical Centre, Leiden, The Netherlands; 2Department of Rheumatology, Leiden University Medical Centre, Leiden, The Netherlands; 3Department of Epidemiology & Biostatistics, Erasmus Medical Centre, Rotterdam, The Netherlands In a genome-wide linkage scan of seven families with familial early-onset osteoarthritis (FOA), we mapped a FOA locus to a 5 cM region on chromosome 2q33.3–2q34 with a maximum LOD score of 6.05. To identify causal variants, 17 positional candidate genes and FRZB were sequenced for coding, splice sites, and 50 and 30 untranslated regions. The pathogenicity of possible disease-causing variants was evaluated using predicted effects on protein structure and function, splicing enhancers, degree of conservation and frequency in 790 unrelated subjects from the population-based Rotterdam study scored for the presence of radiographic signs of OA (ROA). Nine novel variants, identified in NRP2, XM_371590, ADAM23, IDH1, PIP5K3 and PTHR2, cosegregated with FOA, of which two were promising. IDH1 Y183C cosegregated in one family, involved a conserved amino-acid change and showed a damaging effect predicted by PolyPhen and SIFT. In the Rotterdam sample, carriers of IDH1 Y183C (0.02) had an increased but insignificant risk for generalised ROA. The second variant, NRP2 c.1938-21T4C cosegregated in three families. -

Expression of ADAMTS13 and PCNA in the Placentas of Gestational Diabetic Mothers
Int. J. Morphol., 39(1):38-44, 2021. Expression of ADAMTS13 and PCNA in the Placentas of Gestational Diabetic Mothers Expresión de ADAMTS13 y PCNA en las Placentas de Madres Diabéticas Gestacionales Süleyman Cemil Oglak1 & Mehmet Obut1 OGLAK, S. C. & OBUT, M. Expression of ADAMTS13 and PCNA in the placentas of gestational diabetic mothers. Int. J. Morphol., 39(1):38-44, 2021. SUMMARY: GDM is linked with overexpression of inflammatory cytokines and increased oxidative stress, leading to endothelial dysfunction and vascular disorder. Weaimed to examine the expression of ADAMTS13 and PCNA in the placentas of gestational diabe- tes mellitus (GDM) patients to investigate the effects of hypoxia, induced by GDM, on proliferation and extracellular matrix formation in the maternal and fetal placenta cells. A total of 60 placentas were collected from pregnant women admitted to the obstetrics clinic. Thirty of them were diagnosed with GDM, and 30 of them were diagnosed with non-GDM patients. Samples were fixed in 10 % formaldehyde, after routine follow-up, embedded in paraffin wax. Sections of 5 µm were cut stained with Mayer Hematoxylin-Eosin, examined under a light microscope. Sections for immunohistochemical analysis were cut and processed for antigen retrievalin citrate solution. Sections were incubated with ADAMTS13 and PCNA primary antibodies, counterstained with hematoxylin, and evaluate under a light microscope. In histopathological examination, the non-diabetic placentas showed that decidua cells in the maternal region were polygonal with oval nuclei and organized in groups. In the GDM group, there were pyknosis and apoptotic changes in decidua cell nuclei. Vacuolar areas were observed in large cavities in maternal connective tissue.