4182575.Pdf (1014.Kb)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Folding Switch Regulates Interleukin 27 Biogenesis and Secretion of Its Α-Subunit As a Cytokine
A folding switch regulates interleukin 27 biogenesis and secretion of its α-subunit as a cytokine Stephanie I. Müllera,b, Antonie Friedlc, Isabel Aschenbrennera,b, Julia Esser-von Bierenc, Martin Zachariasd, Odile Devergnee,1, and Matthias J. Feigea,b,1 aCenter for Integrated Protein Science, Department of Chemistry, Technical University of Munich, 85748 Garching, Germany; bInstitute for Advanced Study, Technical University of Munich, 85748 Garching, Germany; cCenter of Allergy and Environment (ZAUM), Technical University of Munich and Helmholtz Center Munich, 80802 Munich, Germany; dCenter for Integrated Protein Science, Physics Department, Technical University of Munich, 85748 Garching, Germany; and eSorbonne Université, INSERM, CNRS, Centre d’Immunologie et des Maladies Infectieuses, 75 013 Paris, France Edited by John J. O’Shea, NIH, Bethesda, MD, and accepted by Editorial Board Member Tadatsugu Taniguchi December 10, 2018 (received for review October 3, 2018) A common design principle of heteromeric signaling proteins is the by the secretion and biological activity of some isolated α-and use of shared subunits. This allows encoding of complex messages β-subunits (10, 11). A prominent example is IL-27α/p28. Murine IL- while maintaining evolutionary flexibility. How cells regulate and 27α, also designated as IL-30, is secreted in isolation (12) and per- control assembly of such composite signaling proteins remains an forms immunoregulatory roles (13–16). In contrast, no autonomous important open question. An example of particular complexity and secretion of human IL-27α has been reported yet. The molecular biological relevance is the interleukin 12 (IL-12) family. Four func- basis for this difference has remained unclear, but it is likely to have tionally distinct αβ heterodimers are assembled from only five sub- a profound impact on immune system function, since in mice IL-27 units to regulate immune cell function and development. -

Role of Amylase in Ovarian Cancer Mai Mohamed University of South Florida, [email protected]
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School July 2017 Role of Amylase in Ovarian Cancer Mai Mohamed University of South Florida, [email protected] Follow this and additional works at: http://scholarcommons.usf.edu/etd Part of the Pathology Commons Scholar Commons Citation Mohamed, Mai, "Role of Amylase in Ovarian Cancer" (2017). Graduate Theses and Dissertations. http://scholarcommons.usf.edu/etd/6907 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Role of Amylase in Ovarian Cancer by Mai Mohamed A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Department of Pathology and Cell Biology Morsani College of Medicine University of South Florida Major Professor: Patricia Kruk, Ph.D. Paula C. Bickford, Ph.D. Meera Nanjundan, Ph.D. Marzenna Wiranowska, Ph.D. Lauri Wright, Ph.D. Date of Approval: June 29, 2017 Keywords: ovarian cancer, amylase, computational analyses, glycocalyx, cellular invasion Copyright © 2017, Mai Mohamed Dedication This dissertation is dedicated to my parents, Ahmed and Fatma, who have always stressed the importance of education, and, throughout my education, have been my strongest source of encouragement and support. They always believed in me and I am eternally grateful to them. I would also like to thank my brothers, Mohamed and Hussien, and my sister, Mariam. I would also like to thank my husband, Ahmed. -

Ncounter® Mouse Autoimmune Profiling Panel - Gene and Probe Details
nCounter® Mouse AutoImmune Profiling Panel - Gene and Probe Details Official Symbol Accession Alias / Previous Symbol Official Full Name Other targets or Isoform Information AW208573,CD143,expressed sequence AW208573,MGD-MRK- Ace NM_009598.1 1032,MGI:2144508 angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 2610036I19Rik,2610510L13Rik,Acinus,apoptotic chromatin condensation inducer in the nucleus,C79325,expressed sequence C79325,MGI:1913562,MGI:1919776,MGI:2145862,mKIAA0670,RIKEN cDNA Acin1 NM_001085472.2 2610036I19 gene,RIKEN cDNA 2610510L13 gene apoptotic chromatin condensation inducer 1 Acp5 NM_001102405.1 MGD-MRK-1052,TRACP,TRAP acid phosphatase 5, tartrate resistant 2310066K23Rik,AA960180,AI851923,Arp1b,expressed sequence AA960180,expressed sequence AI851923,MGI:2138136,MGI:2138359,RIKEN Actr1b NM_146107.2 cDNA 2310066K23 gene ARP1 actin-related protein 1B, centractin beta Adam17 NM_001277266.1 CD156b,Tace,tumor necrosis factor-alpha converting enzyme a disintegrin and metallopeptidase domain 17 ADAR1,Adar1p110,Adar1p150,AV242451,expressed sequence Adar NM_001038587.3 AV242451,MGI:2139942,mZaADAR adenosine deaminase, RNA-specific Adora2a NM_009630.2 A2AAR,A2aR,A2a, Rs,AA2AR,MGD-MRK-16163 adenosine A2a receptor Ager NM_007425.2 RAGE advanced glycosylation end product-specific receptor AI265500,angiotensin precursor,Aogen,expressed sequence AI265500,MGD- Agt NM_007428.3 MRK-1192,MGI:2142488,Serpina8 angiotensinogen (serpin peptidase inhibitor, clade A, member 8) Ah,Ahh,Ahre,aromatic hydrocarbon responsiveness,aryl hydrocarbon -

Interleukin-30/Il27p28 Shapes Prostate Cancer Stem-Like Cell Behavior and Is Critical for Tumor Onset and Metastasization Carlo Sorrentino1,2,3, Stefania L
Published OnlineFirst February 27, 2018; DOI: 10.1158/0008-5472.CAN-17-3117 Cancer Tumor Biology and Immunology Research Interleukin-30/IL27p28 Shapes Prostate Cancer Stem-like Cell Behavior and Is Critical for Tumor Onset and Metastasization Carlo Sorrentino1,2,3, Stefania L. Ciummo1,2,3, Giuseppe Cipollone4,5, Sara Caputo6, Matteo Bellone6, and Emma Di Carlo1,2,3 Abstract Prostate cancer stem-like cells (PCSLC) are believed to be signaling. IL30 overproduction by PCSLCs promoted tumor responsible for prostate cancer onset and metastasis. Autocrine onset and development associated with increased proliferation, and microenvironmental signals dictate PCSLC behavior and vascularization, and myeloid cell recruitment. Furthermore, it patient outcome. In prostate cancer patients, IL30/IL27p28 has promoted PCSLC dissemination to lymph nodes and bone been linked with tumor progression, but the mechanisms marrow by upregulating the CXCR5/CXCL13 axis, and drove underlying this link remain mostly elusive. Here, we asked metastasis to lungs through the CXCR4/CXCL12 axis. These whether IL30 may favor prostate cancer progression by condi- mechanisms were drastically hindered by IL30 knockdown tioning PCSLCs and assessed the value of blocking IL30 to or knockout in PCSLCs. Collectively, these results mark IL30 suppress tumor growth. IL30 was produced by PCSLCs in as a key driver of PCSLC behavior. Targeting IL30 signaling may human and murine prostatic intraepithelial neoplasia and be a potential therapeutic strategy against prostate cancer pro- displayed significant autocrine and paracrine effects. PCSLC- gression and recurrence. derived IL30 supported PCSLC viability, self-renewal and Significance: IL30 plays an important role in regulating pro- tumorigenicity, expression of inflammatory mediators and state cancer stem-like cell behavior and metastatic potential, growth factors, tumor immune evasion, and regulated chemo- therefore targeting this cytokine could hamper prostate cancer kine and chemokine receptor genes, primarily via STAT1/STAT3 progression or recurrence. -

Wsx-1/Il-27 As a Target for Anti-Inflammatory Responses
(19) TZZ ZZ_T (11) EP 2 046 809 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C07K 1/00 (2006.01) C07K 14/54 (2006.01) 07.12.2016 Bulletin 2016/49 C07K 14/715 (2006.01) C07K 16/24 (2006.01) (21) Application number: 07836143.3 (86) International application number: PCT/US2007/016329 (22) Date of filing: 18.07.2007 (87) International publication number: WO 2008/011081 (24.01.2008 Gazette 2008/04) (54) WSX-1/IL-27 AS A TARGET FOR ANTI-INFLAMMATORY RESPONSES WSX-1/IL-27 ALS TARGET FÜR ENTZÜNDUNGSHEMMENDE REAKTIONEN WSX-1/IL-27 UTILISÉ COMME CIBLE POUR SUSCITER DES RÉACTIONS ANTI-INFLAMMATOIRES (84) Designated Contracting States: • VILLARINO ALEJANDRO V ET AL: "IL-27 limits AT BE BG CH CY CZ DE DK EE ES FI FR GB GR IL-2 production during Th1 differentiation." HU IE IS IT LI LT LU LV MC MT NL PL PT RO SE JOURNALOF IMMUNOLOGY (BALTIMORE, MD. : SI SK TR 1950) 1 JAN 2006, vol. 176, no. 1, 1 January 2006 Designated Extension States: (2006-01-01), pages 237-247, XP002570061 ISSN: AL BA RS 0022-1767 • KASTELEIN ROBERT A ET AL: "Discovery and (30) Priority: 19.07.2006 US 832213 P biologyof IL-23 andIL-27: relatedbut functionally 11.08.2006 US 837450 P distinct regulators of inflammation." ANNUAL REVIEW OF IMMUNOLOGY 2007, vol. 25, 2007, (43) Date of publication of application: pages 221-242, XP002570062 ISSN: 0732-0582 15.04.2009 Bulletin 2009/16 • SCHELLER J ET AL: "No inhibition of IL-27 signaling by soluble gp130" BIOCHEMICAL AND (73) Proprietor: The Trustees of The University of BIOPHYSICAL RESEARCH COMMUNICATIONS, Pennsylvania ACADEMIC PRESS INC. -

GP130 Cytokines in Breast Cancer and Bone
cancers Review GP130 Cytokines in Breast Cancer and Bone Tolu Omokehinde 1,2 and Rachelle W. Johnson 1,2,3,* 1 Program in Cancer Biology, Vanderbilt University, Nashville, TN 37232, USA; [email protected] 2 Vanderbilt Center for Bone Biology, Department of Medicine, Division of Clinical Pharmacology, Vanderbilt University Medical Center, Nashville, TN 37232, USA 3 Department of Medicine, Division of Clinical Pharmacology, Vanderbilt University Medical Center, Nashville, TN 37232, USA * Correspondence: [email protected]; Tel.: +1-615-875-8965 Received: 14 December 2019; Accepted: 29 January 2020; Published: 31 January 2020 Abstract: Breast cancer cells have a high predilection for skeletal homing, where they may either induce osteolytic bone destruction or enter a latency period in which they remain quiescent. Breast cancer cells produce and encounter autocrine and paracrine cytokine signals in the bone microenvironment, which can influence their behavior in multiple ways. For example, these signals can promote the survival and dormancy of bone-disseminated cancer cells or stimulate proliferation. The interleukin-6 (IL-6) cytokine family, defined by its use of the glycoprotein 130 (gp130) co-receptor, includes interleukin-11 (IL-11), leukemia inhibitory factor (LIF), oncostatin M (OSM), ciliary neurotrophic factor (CNTF), and cardiotrophin-1 (CT-1), among others. These cytokines are known to have overlapping pleiotropic functions in different cell types and are important for cross-talk between bone-resident cells. IL-6 cytokines have also been implicated in the progression and metastasis of breast, prostate, lung, and cervical cancer, highlighting the importance of these cytokines in the tumor–bone microenvironment. This review will describe the role of these cytokines in skeletal remodeling and cancer progression both within and outside of the bone microenvironment. -
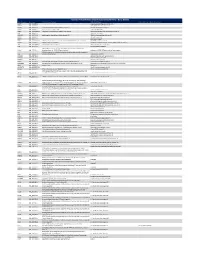
Ncounter® Autoimmune Discovery Consortium Panel
nCounter® Autoimmune Discovery Consortium Panel - Gene Details Official Symbol Accession Alias / Previous Symbol Official Full Name Other targets or Isoform Information AAMP NM_001087.3 angio associated migratory cell protein ABHD6 NM_020676.5 abhydrolase domain containing 6 ACKR2 NM_001296.3 CMKBR9,CCBP2;chemokine binding protein 2 atypical chemokine receptor 2 ACOXL NM_018308.1 acyl-Coenzyme A oxidase-like acyl-CoA oxidase like ACSL6 NM_001009185.1 FACL6;fatty-acid-Coenzyme A ligase, long-chain 6 acyl-CoA synthetase long chain family member 6 ADA NM_000022.2 adenosine deaminase ADAM30 NM_021794.2 a disintegrin and metalloproteinase domain 30 ADAM metallopeptidase domain 30 ADCY3 NM_004036.3 adenylate cyclase 3 ADCY7 NM_001114.4 adenylate cyclase 7 AFF3 NM_001025108.1 LAF4;lymphoid nuclear protein related to AF4,AF4/FMR2 family, member 3 AF4/FMR2 family member 3 AGAP2 NM_014770.3 CENTG1;centaurin, gamma 1 ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 AHI1 NM_001134830.1 Abelson helper integration site Abelson helper integration site 1 AHR NM_001621.3 aryl hydrocarbon receptor AHSA2;AHA1, activator of heat shock 90kDa protein ATPase homolog 2 AHSA2 NM_152392.1 (yeast),activator of HSP90 ATPase homolog 2 activator of HSP90 ATPase homolog 2, pseudogene APECED;autoimmune regulator (autoimmune polyendocrinopathy candidiasis AIRE NM_000383.2 ectodermal dystrophy) autoimmune regulator AMIGO3 NM_198722.2 adhesion molecule with Ig like domain 3 ANKRD55 NM_024669.2 ankyrin repeat domain 55 ANTXR2 NM_058172.5 anthrax toxin receptor -

Advances in Therapeutic Targeting of Cancer Stem Cells Within the Tumor Microenvironment: an Updated Review
cells Review Advances in Therapeutic Targeting of Cancer Stem Cells within the Tumor Microenvironment: An Updated Review 1,2, , 1,2, 1,2, Kevin Dzobo * y , Dimakatso Alice Senthebane y, Chelene Ganz y, Nicholas Ekow Thomford 3,4 , Ambroise Wonkam 3 and Collet Dandara 3 1 International Centre for Genetic Engineering and Biotechnology (ICGEB), Cape Town Component, Wernher and Beit Building (South), UCT Medical Campus, Anzio Road, Observatory, Cape Town 7925, South Africa; [email protected] (D.A.S.); [email protected] (C.G.) 2 Division of Medical Biochemistry and Institute of Infectious Disease and Molecular Medicine, Department of Integrative Biomedical Sciences, Faculty of Health Sciences, University of Cape Town, Cape Town 7925, South Africa 3 Division of Human Genetics, Department of Pathology and Institute for Infectious Disease and Molecular Medicine, Faculty of Health Sciences, University of Cape Town, Anzio Road, Observatory, Cape Town 7925, South Africa; [email protected] (N.E.T.); [email protected] (A.W.); [email protected] (C.D.) 4 Department of Medical Biochemistry, School of Medical Sciences, College of Health Sciences, University of Cape Coast, PMB, Cape Coast, Ghana * Correspondence: [email protected]; Tel.: +27-842953708 These authors contributed equally to this work. y Received: 7 July 2020; Accepted: 11 August 2020; Published: 13 August 2020 Abstract: Despite great strides being achieved in improving cancer patients’ outcomes through better therapies and combinatorial treatment, several hurdles still remain due to therapy resistance, cancer recurrence and metastasis. Drug resistance culminating in relapse continues to be associated with fatal disease. The cancer stem cell theory posits that tumors are driven by specialized cancer cells called cancer stem cells (CSCs). -

C07k - 2021.08
CPC - C07K - 2021.08 C07K PEPTIDES (peptides in foodstuffs A23; obtaining protein compositions for foodstuffs, working-up proteins for foodstuffs A23J; preparations for medicinal purposes A61K; peptides containing beta-lactam rings C07D; cyclic dipeptides not having in their molecule any other peptide link than those which form their ring, e.g. piperazine-2,5-diones, C07D; ergot alkaloids of the cyclic peptide type C07D 519/02; macromolecular compounds having statistically distributed amino acid units in their molecules, i.e. when the preparation does not provide for a specific; but for a random sequence of the amino acid units, homopolyamides and block copolyamides derived from amino acids C08G 69/00; macromolecular products derived from proteins C08H 1/00; preparation of glue or gelatine C09H; single cell proteins, enzymes C12N; genetic engineering processes for obtaining peptides C12N 15/00; compositions for measuring or testing processes involving enzymes C12Q; investigation or analysis of biological material G01N 33/00) Relationships with other classification places An amino acid per se is classified in C07D while peptides (starting from dipeptides) are classified in C07K. Subclass C07K is a function oriented entry for the compounds themselves and does not cover the application or use of the compounds under the subclass definition. For classifying such information other entries exist, for example: preservation of bodies of humans or animals or plants or parts thereof; Biocides, e.g. as disinfectants, as pesticides, as herbicides; pest repellants or attractants; plant growth regulators are classified in A01N. Preparations for medical, dental, or toilet purposes are classified in A61K. Amino acids or derivatives thereof are classified in C07C or C07D. -

( 12 ) Patent Application Publication ( 10 ) Pub . No .: US 2019/0321403 A1
INDUS 20190321403A1 IN (19 ) United States (12 ) Patent Application Publication ( 10 ) Pub . No .: US 2019/0321403 A1 LEVITSKY (43 ) Pub . Date : Oct. 24 , 2019 ( 54 ) ENGINEERED B CELLS AND RELATED Publication Classification COMPOSITIONS AND METHODS ( 51 ) Int. Ci. A61K 35/17 (2006.01 ) (71 ) Applicant: Juno Therapeutics , Inc., Seattle, WA CI2N 15/85 (2006.01 ) (US ) C12N 15/62 ( 2006.01 ) A61 ) 1/10 (2006.01 ) ( 72 ) Inventor : Hyam I. LEVITSKY , Seattle , WA C07K 16/28 ( 2006.01 ) (US ) (52 ) U.S. CI. CPC A61K 35/17 (2013.01 ) ; C12N 15/85 ( 73 ) Assignee: Juno Therapeutics, Inc. , Seattle , WA ( 2013.01) ; C12N 15/62 (2013.01 ) ; A61K 45/06 (US ) ( 2013.01 ) ; CO7K 16/28 ( 2013.01) ; C12N 2015/8518 ( 2013.01 ) ; A61J 1/10 ( 2013.01) (21 ) Appl. No .: 16 /465,109 (57 ) ABSTRACT Provided herein are engineered B cells , such as for adoptive ( 22 ) PCT Filed : Nov. 30 , 2017 cell therapy. In some aspects , also provided are methods and compositions for engineering and producing the cells , com ( 86 ) PCT No.: PCT /US2017 / 064075 positions containing the cells , and methods for their admin $ 371 ( c ) ( 1 ) , istration to subjects . In some embodiments , the cells are (2 ) Date : May 29 , 2019 engineered to produce and /or secrete an exogenous protein , such as a therapeutic protein , including antibodies and antigen - binding fragments thereof. In some aspects , features Related U.S. Application Data of the cells and methods provide for increased or improved (60 ) Provisional application No. 62/ 429,709, filed on Dec. activity , efficacy and /or persistence of the cells . 2 , 2016 . -

IL-30† (IL-27A): a Familiar Stranger in Immunity, Inflammation, and Cancer
Min et al. Experimental & Molecular Medicine (2021) 53:823–834 https://doi.org/10.1038/s12276-021-00630-x Experimental & Molecular Medicine REVIEW ARTICLE Open Access IL-30† (IL-27A): a familiar stranger in immunity, inflammation, and cancer Booki Min 1,2,DongkyunKim1 and Matthias J. Feige3 Abstract Over the years, interleukin (IL)-27 has received much attention because of its highly divergent, sometimes even opposing, functions in immunity. IL-30, the p28 subunit that forms IL-27 together with Ebi3 and is also known as IL- 27p28 or IL-27A, has been considered a surrogate to represent IL-27. However, it was later discovered that IL-30 can form complexes with other protein subunits, potentially leading to overlapping or discrete functions. Furthermore, there is emerging evidence that IL-30 itself may perform immunomodulatory functions independent of Ebi3 or other binding partners and that IL-30 production is strongly associated with certain cancers in humans. In this review, we will discuss the biology of IL-30 and other IL-30-associated cytokines and their functions in inflammation and cancer. Introduction extracellular bacterial or fungal infections3,4. The essential Adaptive T cell responses are shaped by multiple fac- protective immunity mediated by these cytokines requires tors, and soluble mediators, especially cytokines produced tight regulation, and inadequate control results in auto- by ‘innate’ immune cells, play an instructive role in the immune inflammation. Targeting or utilizing these key 1 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; process . Heterodimeric cytokines that belong to the IL- cytokines and/or their signaling pathways is being 12 family are instrumental to the generation of inflam- exploited in therapeutic approaches to intervene in matory Th1 and Th17 immunity. -

Human Cytokine Response Profiles
Comprehensive Understanding of the Human Cytokine Response Profiles A. Background The current project aims to collect datasets profiling gene expression patterns of human cytokine treatment response from the NCBI GEO and EBI ArrayExpress databases. The Framework for Data Curation already hosted a list of candidate datasets. You will read the study design and sample annotations to select the relevant datasets and label the sample conditions to enable automatic analysis. If you want to build a new data collection project for your topic of interest instead of working on our existing cytokine project, please read section D. We will explain the cytokine project’s configurations to give you an example on creating your curation task. A.1. Cytokine Cytokines are a broad category of small proteins mediating cell signaling. Many cell types can release cytokines and receive cytokines from other producers through receptors on the cell surface. Despite some overlap in the literature terminology, we exclude chemokines, hormones, or growth factors, which are also essential cell signaling molecules. Meanwhile, we count two cytokines in the same family as the same if they share the same receptors. In this project, we will focus on the following families and use the member symbols as standard names (Table 1). Family Members (use these symbols as standard cytokine names) Colony-stimulating factor GCSF, GMCSF, MCSF Interferon IFNA, IFNB, IFNG Interleukin IL1, IL1RA, IL2, IL3, IL4, IL5, IL6, IL7, IL9, IL10, IL11, IL12, IL13, IL15, IL16, IL17, IL18, IL19, IL20, IL21, IL22, IL23, IL24, IL25, IL26, IL27, IL28, IL29, IL30, IL31, IL32, IL33, IL34, IL35, IL36, IL36RA, IL37, TSLP, LIF, OSM Tumor necrosis factor TNFA, LTA, LTB, CD40L, FASL, CD27L, CD30L, 41BBL, TRAIL, OPGL, APRIL, LIGHT, TWEAK, BAFF Unassigned TGFB, MIF Table 1.