Exploring Pharmacological Space
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Drug Class Review Beta Adrenergic Blockers
Drug Class Review Beta Adrenergic Blockers Final Report Update 4 July 2009 Update 3: September 2007 Update 2: May 2005 Update 1: September 2004 Original Report: September 2003 The literature on this topic is scanned periodically. The purpose of this report is to make available information regarding the comparative effectiveness and safety profiles of different drugs within pharmaceutical classes. Reports are not usage guidelines, nor should they be read as an endorsement of, or recommendation for, any particular drug, use, or approach. Oregon Health & Science University does not recommend or endorse any guideline or recommendation developed by users of these reports. Mark Helfand, MD, MPH Kim Peterson, MS Vivian Christensen, PhD Tracy Dana, MLS Sujata Thakurta, MPA:HA Drug Effectiveness Review Project Marian McDonagh, PharmD, Principal Investigator Oregon Evidence-based Practice Center Mark Helfand, MD, MPH, Director Oregon Health & Science University Copyright © 2009 by Oregon Health & Science University Portland, Oregon 97239. All rights reserved. Final Report Update 4 Drug Effectiveness Review Project TABLE OF CONTENTS INTRODUCTION .......................................................................................................................... 6 Purpose and Limitations of Evidence Reports........................................................................................ 8 Scope and Key Questions .................................................................................................................... 10 METHODS................................................................................................................................. -

Different Effects of Propranolol, Bisoprolol, Carvedilol and Doxazosin on Heart Rate, Blood Pressure, and Plasma Concentrations of Epinephrine and Norepinephrine K
Journal of Clinical and Basic Cardiology An Independent International Scientific Journal Journal of Clinical and Basic Cardiology 2003; 6 (1-4), 69-72 Different Effects of Propranolol Bisoprolol, Carvedilol and Doxazosin on Heart Rate, Blood Pressure, and Plasma Concentrations of Epinephrine and Norepinephrine Stoschitzky K, Donnerer J, Klein W, Koshucharova G Kraxner W, Lercher P, Maier R, Watzinger N, Zweiker R Homepage: www.kup.at/jcbc Online Data Base Search for Authors and Keywords Indexed in Chemical Abstracts EMBASE/Excerpta Medica Krause & Pachernegg GmbH · VERLAG für MEDIZIN und WIRTSCHAFT · A-3003 Gablitz/Austria ORIGINAL PAPERS, CLINICAL CARDIOLOGY Alpha- Versus Beta-Blockers J Clin Basic Cardiol 2003; 6: 69 Different Effects of Propranolol, Bisoprolol, Carvedilol and Doxazosin on Heart Rate, Blood Pressure, and Plasma Concentrations of Epinephrine and Norepinephrine K. Stoschitzky1, G. Koshucharova1, R. Zweiker1, P. Lercher1, R. Maier1, N. Watzinger1, W. Kraxner1, W. Klein1, J. Donnerer2 Background: Despite of its beta-blocking effects, carvedilol has been shown not to decrease resting heart rate in healthy subjects. Therefore, we compared haemodynamic effects of carvedilol (an alpha- and beta-blocker), propranolol (a non-selec- tive beta-blocker), bisoprolol (a beta1-selective beta-blocker), doxazosin (an alpha-blocker) and placebo, at rest and during exercise. In addition, we measured plasma levels of epinephrine and norepinephrine. Methods: Twelve healthy males received single oral doses of 80 mg propranolol, 5 mg bisoprolol, 50 mg carvedilol, 4 mg doxazosin and placebo according to a randomized, double-blind, crossover protocol. Three hours after drug intake, heart rate and blood pressure were measured at rest, after 10 min of exercise, and after 15 min of recovery. -

Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults
PREVENTION, DETECTION, EVALUATION, AND MANAGEMENT OF HIGH BLOOD PRESSURE IN ADULTS Thomas F. Whayne, Jr, MD, PhD, FACC, FAHA, FACP Professor of Medicine (Cardiology) University of Kentucky Gill Heart Institute NOVEMBER 2018 No conflicts to disclose Email: [email protected] (0 number zero) (0 is the number zero) Educational Need/Practice Gap Regarding hypertension, the major practice gap is failure to adequately control BP. Tight BP control as in JNC-7 is less with unofficial JNC-8 (2013) but nevertheless, the need for adequate treatment must still be emphasized. Now, the 2015 NIH study appears more like JNC-7, followed by additional guidelines, all adding to the confusion. Good judgment in clinical practice is still the order of the day. Objectives • Review the latest guidelines and evidence- based medicine for the treatment of hypertension. • Discuss specific mechanisms involved in the causation of hypertension and how this affects treatment. • Summarize specific invasive techniques for the difficult patient whose hypertension does not respond to medications. Expected Outcome Approximately 75% of hypertensive patients are not adequately managed. This is in part the fault of many patients but also there is failure of clinicians to push for maximal beneficial results. Hopefully, a consideration of available information and medications will emphasize the possibilities for improved BP control. Hypertension pts. (50 million: 24% of US population): Treatment and Control % of Hypertensives % on Rx Achieving taking Medications BP Control 24% Controlled on Rx Not 53% Not on Rx on Rx on Rx 29% Not Controlled on Rx NHANES III, The Centers for Disease Control and Prevention, and the National Center for Health Statistics, Burt et. -

Drugs for Primary Prevention of Atherosclerotic Cardiovascular Disease: an Overview of Systematic Reviews
Supplementary Online Content Karmali KN, Lloyd-Jones DM, Berendsen MA, et al. Drugs for primary prevention of atherosclerotic cardiovascular disease: an overview of systematic reviews. JAMA Cardiol. Published online April 27, 2016. doi:10.1001/jamacardio.2016.0218. eAppendix 1. Search Documentation Details eAppendix 2. Background, Methods, and Results of Systematic Review of Combination Drug Therapy to Evaluate for Potential Interaction of Effects eAppendix 3. PRISMA Flow Charts for Each Drug Class and Detailed Systematic Review Characteristics and Summary of Included Systematic Reviews and Meta-analyses eAppendix 4. List of Excluded Studies and Reasons for Exclusion This supplementary material has been provided by the authors to give readers additional information about their work. © 2016 American Medical Association. All rights reserved. 1 Downloaded From: https://jamanetwork.com/ on 09/28/2021 eAppendix 1. Search Documentation Details. Database Organizing body Purpose Pros Cons Cochrane Cochrane Library in Database of all available -Curated by the Cochrane -Content is limited to Database of the United Kingdom systematic reviews and Collaboration reviews completed Systematic (UK) protocols published by by the Cochrane Reviews the Cochrane -Only systematic reviews Collaboration Collaboration and systematic review protocols Database of National Health Collection of structured -Curated by Centre for -Only provides Abstracts of Services (NHS) abstracts and Reviews and Dissemination structured abstracts Reviews of Centre for Reviews bibliographic -

L:\0901 with Peru\0901PHARMAPPX.Wpd
Harmonized Tariff Schedule of the United States (2009) (Rev. 1) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2009) (Rev. 1) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 2 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. ABACAVIR 136470-78-5 ACEXAMIC ACID 57-08-9 ABAFUNGIN 129639-79-8 ACICLOVIR 59277-89-3 ABAMECTIN 65195-55-3 ACIFRAN 72420-38-3 ABANOQUIL 90402-40-7 ACIPIMOX 51037-30-0 ABAPERIDONE 183849-43-6 ACITAZANOLAST 114607-46-4 ABARELIX 183552-38-7 ACITEMATE 101197-99-3 ABATACEPT 332348-12-6 ACITRETIN 55079-83-9 ABCIXIMAB 143653-53-6 ACIVICIN 42228-92-2 ABECARNIL 111841-85-1 ACLANTATE 39633-62-0 ABETIMUS 167362-48-3 ACLARUBICIN 57576-44-0 ABIRATERONE 154229-19-3 ACLATONIUM NAPADISILATE 55077-30-0 ABITESARTAN 137882-98-5 ACODAZOLE 79152-85-5 ABLUKAST 96566-25-5 ACOLBIFENE 182167-02-8 ABRINEURIN 178535-93-8 ACONIAZIDE 13410-86-1 ABUNIDAZOLE 91017-58-2 ACOTIAMIDE 185106-16-5 ACADESINE 2627-69-2 ACOXATRINE 748-44-7 ACAMPROSATE 77337-76-9 ACREOZAST 123548-56-1 ACAPRAZINE 55485-20-6 -

Systematic Evidence Review from the Blood Pressure Expert Panel, 2013
Managing Blood Pressure in Adults Systematic Evidence Review From the Blood Pressure Expert Panel, 2013 Contents Foreword ............................................................................................................................................ vi Blood Pressure Expert Panel ..............................................................................................................vii Section 1: Background and Description of the NHLBI Cardiovascular Risk Reduction Project ............ 1 A. Background .............................................................................................................................. 1 Section 2: Process and Methods Overview ......................................................................................... 3 A. Evidence-Based Approach ....................................................................................................... 3 i. Overview of the Evidence-Based Methodology ................................................................. 3 ii. System for Grading the Body of Evidence ......................................................................... 4 iii. Peer-Review Process ....................................................................................................... 5 B. Critical Question–Based Approach ........................................................................................... 5 i. How the Questions Were Selected ................................................................................... 5 ii. Rationale for the Questions -

Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DIX to the HTSUS—Continued
20558 Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DEPARMENT OF THE TREASURY Services, U.S. Customs Service, 1301 TABLE 1.ÐPHARMACEUTICAL APPEN- Constitution Avenue NW, Washington, DIX TO THE HTSUSÐContinued Customs Service D.C. 20229 at (202) 927±1060. CAS No. Pharmaceutical [T.D. 95±33] Dated: April 14, 1995. 52±78±8 ..................... NORETHANDROLONE. A. W. Tennant, 52±86±8 ..................... HALOPERIDOL. Pharmaceutical Tables 1 and 3 of the Director, Office of Laboratories and Scientific 52±88±0 ..................... ATROPINE METHONITRATE. HTSUS 52±90±4 ..................... CYSTEINE. Services. 53±03±2 ..................... PREDNISONE. 53±06±5 ..................... CORTISONE. AGENCY: Customs Service, Department TABLE 1.ÐPHARMACEUTICAL 53±10±1 ..................... HYDROXYDIONE SODIUM SUCCI- of the Treasury. NATE. APPENDIX TO THE HTSUS 53±16±7 ..................... ESTRONE. ACTION: Listing of the products found in 53±18±9 ..................... BIETASERPINE. Table 1 and Table 3 of the CAS No. Pharmaceutical 53±19±0 ..................... MITOTANE. 53±31±6 ..................... MEDIBAZINE. Pharmaceutical Appendix to the N/A ............................. ACTAGARDIN. 53±33±8 ..................... PARAMETHASONE. Harmonized Tariff Schedule of the N/A ............................. ARDACIN. 53±34±9 ..................... FLUPREDNISOLONE. N/A ............................. BICIROMAB. 53±39±4 ..................... OXANDROLONE. United States of America in Chemical N/A ............................. CELUCLORAL. 53±43±0 -
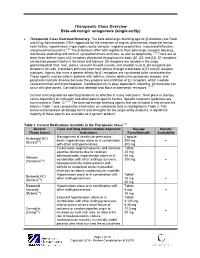
Therapeutic Class Overview Beta-Adrenergic Antagonists (Single-Entity)
Therapeutic Class Overview Beta-adrenergic antagonists (single-entity) · Therapeutic Class Overview/Summary: The beta-adrenergic blocking agents (β-blockers) are Food and Drug Administration (FDA)-approved for the treatment of angina, arrhythmias, essential tremor, heart failure, hypertension, hypertrophic aortic stenosis, migraine prophylaxis, myocardial infarction, and pheochromocytoma.1-26 The β-blockers differ with regards to their adrenergic-receptor blocking, membrane stabilizing and intrinsic sympathomimetic activities, as well as lipophilicity.1-26 There are at least three distinct types of β receptors distributed throughout the body (β1, β2, and β3). β1-receptors are located predominantly in the heart and kidneys. β2-receptors are located in the lungs, gastrointestinal tract, liver, uterus, vascular smooth muscle, and skeletal muscle. β3-receptors are located in fat cells. β-blockers primarily exert their effects through a blockade of β1 and β2 receptor subtypes. Agents that have a greater affinity for β1 receptors are considered to be cardioselective. These agents may be safer in patients with asthma, chronic obstructive pulmonary disease, and peripheral vascular disease because they produce less inhibition of β2 receptors, which mediate vasoconstriction and bronchospasm. Cardioselectivity is dose dependent; therefore, β2 blockade can occur at higher doses. Carvedilol and labetalol also block α-adrenergic receptors. 27-28 Current clinical guidelines identify β-blockers as effective in many indications. Their place in therapy varies depending on indication and other patient specific factors. Specific treatment guidelines are summarized in Table 12.29-61 The beta-adrenergic blocking agents that are included in this review are listed in Table 1 and comparative information on cardioselectivity is highlighted in Table 2. -

Pharmacology of Resistant Hypertension
7/25/2013 47th Annual Meeting ҉ August 2-4, 2013 ҉ Orlando, FL Presenter Disclosure Information Financial Disclosure: I do not have a vested interest in or affiliation with any Pharmacology of corporate organization offering financial support or grant monies for this continuing education activity, or any affiliation with an organization whose Resistant Hypertension philosophy could potentially bias my presentation. Other Disclosures: I have participated in the development of the “PRA and David Parra, Pharm.D., FCCP, BCPS HTN” app for the iPhone and Android smartphones. The app guides the user Clinical Pharmacy Specialist, Cardiology in the use of plasma renin activity to guide anti‐hypertensive therapy. I West Palm Beach VAMC received no compensation for its development or for its use which is available for download free of charge. Clinical Associate Professor Department of Experimental and Clinical Pharmacology College of Pharmacy, University of Minnesota The views expressed in this presentation reflect those of the author, and not necessarily those of the Department of Veterans Affairs. Objectives Consequences of Uncontrolled Hypertension •Describe the general approach to the drug management of resistant hypertension •Recognize commonly prescribed medications that may impede blood pressure control •Compare and contrast the pharmacokinetics/ancillary properties of antihypertensive agents to select the most appropriate agent when presented with a patient case • Apply the concept of plasma renin activity to help guide treatment decisions with antihypertensive agents Messerli F. N Engl J Med 1995;332:1038-1039 Prevalence, Treatment, and Headlines of the St. Louis Post-Dispatch, April 13, 1945 Control of Hypertension 1999–2002 & 2005–2008 Percent of U.S. -

Beta-Blockers for Heart Failure: HT Ong Heart Clinic, Penang, Malaysia (Dr
CASE 1 c Hean T. Ong, FRCP, FACC, FESC; Fei P. Kow, MBBS, MMed Beta-blockers for heart failure: HT Ong Heart Clinic, Penang, Malaysia (Dr. Ong); BBAI Government Health Clinic, Why you should use them more Penang (Dr. Kow) [email protected] Many physicians are afraid to prescribe beta-blockers for patients with heart failure. Yet in most cases, not The authors reported no potential conflict of interest relevant to this article. prescribing them is a mistake. he evidence is clear: Beta-blockers reduce mortality Practice and hospitalization in patients with systolic heart fail- recommendatiOnS Ture.1-3 Yet this class of drugs is underutilized by phy- › Initiate beta-blocker sicians who fear that beta-blocker’s negative inotropic effect therapy in low doses for will lead to worsening heart failure.4 patients with heart fail- Our aim in presenting this review is to counter such con- ure, and increase the dose cerns by detailing the latest evidence. We draw on current gradually until the target research findings to answer questions about beta-blocker dosage is achieved. A selection and dosage and address common misconceptions. › The benefit of beta-blocker therapy for patients with heart failure is propor- Do beta-blockers lower mortality rates tional to the degree of for patients with heart failure? heart rate reduction. A Yes. Three beta-blockers—bisoprolol, carvedilol, and meto- › Consider beta-blocker prolol succinate—have been conclusively shown to reduce therapy for patients with morbidity as well as mortality in patients with systolic heart coexisting chronic obstruc- failure (TABLE 1).1-3,5,6 Here’s a look at the studies: tive pulmonary disease z Bisoprolol. -

Stembook 2018.Pdf
The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances FORMER DOCUMENT NUMBER: WHO/PHARM S/NOM 15 WHO/EMP/RHT/TSN/2018.1 © World Health Organization 2018 Some rights reserved. This work is available under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 IGO licence (CC BY-NC-SA 3.0 IGO; https://creativecommons.org/licenses/by-nc-sa/3.0/igo). Under the terms of this licence, you may copy, redistribute and adapt the work for non-commercial purposes, provided the work is appropriately cited, as indicated below. In any use of this work, there should be no suggestion that WHO endorses any specific organization, products or services. The use of the WHO logo is not permitted. If you adapt the work, then you must license your work under the same or equivalent Creative Commons licence. If you create a translation of this work, you should add the following disclaimer along with the suggested citation: “This translation was not created by the World Health Organization (WHO). WHO is not responsible for the content or accuracy of this translation. The original English edition shall be the binding and authentic edition”. Any mediation relating to disputes arising under the licence shall be conducted in accordance with the mediation rules of the World Intellectual Property Organization. Suggested citation. The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances. Geneva: World Health Organization; 2018 (WHO/EMP/RHT/TSN/2018.1). Licence: CC BY-NC-SA 3.0 IGO. Cataloguing-in-Publication (CIP) data. -

Bucindolol Reduced Mortality and Hospitalisation Related to Cardiovascular Causes in Advanced Chronic Heart Failure
Evid Based Med: first published as 10.1136/ebm.6.6.173 on 1 November 2001. Downloaded from Bucindolol reduced mortality and hospitalisation related to cardiovascular causes in advanced chronic heart failure The Beta-Blocker Evaluation of Survival Trial Investigators. A trial of the beta-blocker bucindolol in patients with advanced chronic heart failure. N Engl J Med 2001 May 31;344:1659–67. QUESTION: In patients with advanced chronic heart failure, does bucindolol reduce all-cause mortality, cardiovascular mortality, and hospitalisation for chronic heart failure? Design Intervention Randomised (allocation concealed*), blinded (clinicians Patients were allocated to bucindolol, 3 mg twice daily and patients),* placebo controlled trial with mean follow for 1 week, which was then titrated gradually to a maxi- up of 2 years (Beta-Blocker Evaluation of Survival Trial mum dose of 100 mg twice daily (n = 1354) or to [BEST]). placebo (n = 1354). Sources of funding: US National Heart, Lung, Main outcome measures and Blood Institute; Setting All-cause mortality, cardiovascular mortality, and hospi- Department of Veterans 90 clinical sites in the USA and Canada. talisation related to chronic heart failure. Affairs Cooperative Studies Program; Incara Main results Pharmaceuticals Patients Analysis was by intention to treat. The groups did not (drugs). 2708 patients (mean age 60 y, 78% men). Inclusion cri- differ for all-cause mortality (adjusted p = 0.13) (table). Patients in the bucindolol group had a lower rate of For correspondence: teria were New York Heart Association (NYHA) class III Dr E J Eichhorn, or IV chronic heart failure caused by primary or cardiovascular mortality (p = 0.04) and hospitalisation Cardiac secondary dilated cardiomyopathy; left ventricular ejec- for chronic heart failure (p < 0.001) than did patients in Catheterization tion fraction < 35%; optimal medical treatment, includ- the placebo group (table).