Impacts of Forest Disturbance on Small Mammal Distribution Allyson Lenora Degrassi University of Vermont
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Woodland Vole Microtus Pinetorum
woodland vole Microtus pinetorum Kingdom: Animalia FEATURES Phylum: Chordata The woodland vole has red-brown body fur with Class: Mammalia light red-brown fur on the belly. The head-body Order: Rodentia length is about three to four inches. The claws on the front feet are larger than the ones on the back Family: Cricetidae feet. The tail is very short, and the eyes are tiny. ILLINOIS STATUS common, native BEHAVIORS The woodland vole may be found statewide in Illinois. This rodent lives on the forest floor in dry woods with oaks, hickories and maples. It also lives in roadside vegetation, orchards, pastures and weedy fields. The woodland vole eats berries, roots, nuts, seeds, wild onions and a variety of green vegetation. It is active day and night. This vole uses burrows under the soil that it digs or that were dug by other small mammals. It digs with the front feet, using the hind feet to push the loose soil behind it. representative specimen The head is used to push loose dirt out of the burrow. The nest is built in a burrow or under an object on the ground that can be reached by a branch of the burrow. Several woodland voles may use the same nest. Two mating seasons occur in a year with young born from March through April and August through November. A litter contains two or three young. Young are helpless at birth but develop rapidly, living on their own at about three weeks of ILLINOIS RANGE age. Sexual maturity is attained between two and three months of age. -

Rock Vole (Microtus Chrotorrhinus)
Rock Vole (Microtus chrotorrhinus) RANGE: Cape Breton Island and e. Quebec w. tone. Min- HOMERANGE: Unknown. nesota. The mountains of n. New England, s, in the Ap- palachians to North Carolina. FOOD HABITS:Bunchberry, wavy-leafed thread moss, blackberry seeds (Martin 1971). May browse on RELATIVEABUNDANCE IN NEW ENGLAND:Unknown, possi- blueberry bushes (twigs and leaves), mushrooms, and bly rare, but may be locally common in appropriate hab- Clinton's lily. A captive subadult ate insects (Timm et al. itat. 1977). Seems to be diurnal with greatest feeding activity taking place in morning (Martin 1971). Less active in HABITAT:Coniferous and mixed forests at higher eleva- afternoon in northern Minnesota (Timm et al. 1977). tions. Favors cool, damp, moss-covered rocks and talus slopes in vicinity of streams. Kirkland (1977a) captured COMMENTS:Occurs locally in small colonies throughout rock voles in clearcuts in West Virginia, habitat not pre- its range. Natural history information is lacking for this viously reported for this species. Timm and others (1977) species. Habitat preferences seem to vary geographi- found voles using edge between boulder field and ma- cally. ture forest in Minnesota. They have been taken at a new low elevation (1,509 feet, 460 m) in the Adirondacks KEY REFERENCES:Banfield 1974, Burt 1957, Kirkland (Kirkland and Knipe 1979). l97?a, Martin 1971, Timm et al. 1977. SPECIALHABITAT REQUIREMENTS: Cool, moist, rocky woodlands with herbaceous groundcover and flowing water. REPRODUCTION: Age at sexual maturity: Females and males are mature when body length exceeds 140 mm and 150 mm, respectively, and total body weight exceeds 30 g for both sexes (Martin 1971). -

Diverzita a Biologie Kryptosporidií Hrabošovitých (Arvicolinae)
JIHOČESKÁ UNIVERZITA V ČESKÝCH BUDĚJOVICÍCH ZEMĚDĚLSKÁ FAKULTA Diverzita a biologie kryptosporidií hrabošovitých (Arvicolinae) Diversity and biology of Cryptosporidium in Arvicolinae rodents disertační práce Ing. Michaela Horčičková Školitel: prof. Ing. Martin Kváč, Ph.D. České Budějovice, 2018 Disertační práce Horčičková, M. 2018: Diverzita a biologie kryptosporidií hrabošovitých (Arvicolinae) [Diversity and biology of Cryptosporidium in Arvicolinae rodents]. Jihočeská univerzita v Českých Budějovicích, Zemědělská fakulta, 123 s. PROHLÁŠENÍ Předkládám tímto k posouzení a obhajobě disertační práci zpracovanou na závěr doktorského studia na Zemědělské fakultě Jihočeské univerzity v Českých Budějovicích. Prohlašuji tímto, že jsem práci vypracovala samostatně, s použitím odborné literatury a dostupných zdrojů uvedených v seznamu, jenž je součástí této práce. Dále prohlašuji, že v souladu s § 47b zákona č. 111/1998 Sb. v platném znění, souhlasím se zveřejněním své disertační práce a to v úpravě vzniklé vypuštěním vyznačených částí archivovaných Zemědělskou fakultou, elektronickou cestou ve veřejně přístupné sekci databáze STAG, provozované Jihočeskou univerzitou v Českých Budějovicích na jejích internetových stránkách. Prohlášení o vědeckém příspěvku výsledků práce Tato disertační práce je založena na výsledcích řady vědeckých publikací, které vznikly za účasti dalších spoluautorů. Na tomto místě prohlašuji, že jsem v rámci studia diverzity a hostitelské specifity kryptosporidií parazitujících u hrabošovitých provedla většinu původního výzkumu a tato práce je založena na vědeckých výsledcích, jimiž jsem hlavní autorkou. V Českých Budějovicích dne 8. července 2018 …………………………… Ing. Michaela Horčičková SEZNAM IMPAKTOVANÝCH PUBLIKACÍ Disertační práce vychází z těchto publikací: Horčičková M., Čondlová Š., Holubová N., Sak B., Květoňová D., Hlásková L., Konečný R., Sedláček F., Clark M.E., Giddings C., McEvoy J.M., Kváč M. 2018: Diversity of Cryptosporidium in common voles and description of Cryptosporidium alticolis sp. -

Investigating Evolutionary Processes Using Ancient and Historical DNA of Rodent Species
Investigating evolutionary processes using ancient and historical DNA of rodent species Thesis submitted for the degree of Doctor of Philosophy (PhD) University of London Royal Holloway University of London Egham, Surrey TW20 OEX Selina Brace November 2010 1 Declaration I, Selina Brace, declare that this thesis and the work presented in it is entirely my own. Where I have consulted the work of others, it is always clearly stated. Selina Brace Ian Barnes 2 “Why should we look to the past? ……Because there is nowhere else to look.” James Burke 3 Abstract The Late Quaternary has been a period of significant change for terrestrial mammals, including episodes of extinction, population sub-division and colonisation. Studying this period provides a means to improve understanding of evolutionary mechanisms, and to determine processes that have led to current distributions. For large mammals, recent work has demonstrated the utility of ancient DNA in understanding demographic change and phylogenetic relationships, largely through well-preserved specimens from permafrost and deep cave deposits. In contrast, much less ancient DNA work has been conducted on small mammals. This project focuses on the development of ancient mitochondrial DNA datasets to explore the utility of rodent ancient DNA analysis. Two studies in Europe investigate population change over millennial timescales. Arctic collared lemming (Dicrostonyx torquatus) specimens are chronologically sampled from a single cave locality, Trou Al’Wesse (Belgian Ardennes). Two end Pleistocene population extinction-recolonisation events are identified and correspond temporally with - localised disappearance of the woolly mammoth (Mammuthus primigenius). A second study examines postglacial histories of European water voles (Arvicola), revealing two temporally distinct colonisation events in the UK. -

Woodland Vole
Woodland Vole Microtus pinetorum Taxa: Mammalian SE-GAP Spp Code: mWOVO Order: Rodentia ITIS Species Code: 180314 Family: Cricetidae NatureServe Element Code: AMAFF11150 KNOWN RANGE: PREDICTED HABITAT: P:\Proj1\SEGap P:\Proj1\SEGap Range Map Link: http://www.basic.ncsu.edu/segap/datazip/maps/SE_Range_mWOVO.pdf Predicted Habitat Map Link: http://www.basic.ncsu.edu/segap/datazip/maps/SE_Dist_mWOVO.pdf GAP Online Tool Link: http://www.gapserve.ncsu.edu/segap/segap/index2.php?species=mWOVO Data Download: http://www.basic.ncsu.edu/segap/datazip/region/vert/mWOVO_se00.zip PROTECTION STATUS: Reported on March 14, 2011 Federal Status: --- State Status: KY (N), MI (SC), MN (SPC), MS (Non-game species in need of management), NJ (S), NY (U), RI (Not Listed), WI (SC/N), ON (SC), QC (Susceptible) NS Global Rank: G5 NS State Rank: AL (S5), AR (S5), CT (S5), DC (S4), DE (S4), FL (SNR), GA (S5), IA (S3), IL (S5), IN (S4), KS (S5), KY (S5), LA (S4), MA (S5), MD (S5), ME (S1), MI (S3S4), MN (S3), MO (SNR), MS (S5), NC (S5), NE (S1), NH (S4), NJ (S4), NY (S5), OH (SNR), OK (S5), PA (S5), RI (SU), SC (SNR), TN (S5), TX (S3), VA (S5), VT (S3), WI (S1), WV (S4), ON (S3?), QC (S3) mWOVO Page 1 of 5 SUMMARY OF PREDICTED HABITAT BY MANAGMENT AND GAP PROTECTION STATUS: US FWS US Forest Service Tenn. Valley Author. US DOD/ACOE ha % ha % ha % ha % Status 1 16,203.8 < 1 30,332.9 < 1 0.0 0 0.0 0 Status 2 40,799.8 < 1 345,823.0 < 1 0.0 0 2,884.8 < 1 Status 3 2,515.1 < 1 1,909,853.9 4 47,162.0 < 1 224,874.2 < 1 Status 4 52.0 < 1 0.0 0 0.0 0 24.8 < 1 Total 59,570.6 < 1 2,286,009.8 5 47,162.0 < 1 227,783.7 < 1 US Dept. -

Virginia Journal of Science Official Publication of the Virginia Academy of Science
VIRGINIA JOURNAL OF SCIENCE OFFICIAL PUBLICATION OF THE VIRGINIA ACADEMY OF SCIENCE Vol. 60 No. 2 Summer 2009 TABLE OF CONTENTS ARTICLES PAGE ABSTRACTS OF PAPERS, 87th Annual Meeting of the Virginia Academy of Science, May 27-29, 2009, Virginia Commonwealth University, Richmond, VA SECTION ABSTRACTS Aeronautical and Aerospace Sciences 53 Agriculture, Forestry and Aquaculture Science 55 Astronomy, Mathematics and Physics &Materials Science 61 Biology and Microbiology & Molecular Biology 64 Biomedical and General Engineering 72 Botany 72 Chemistry 76 Computer Science 83 Education 84 Environmental Science 86 Medical Science 91 Natural History & Biodiversity 98 Psychology 102 Statistics 107 BEST STUDENT PAPER AWARDS 109 JUNIOR ACADEMY AWARDS 113 NEW FELLOWS 127 AUTHOR INDEX 133 ABSTRACTS OF PAPERS, 87th Annual Meeting of the Virginia Academy of Science, May 27-29, 2009, Virginia Commonwealth University, Richmond VA Aeronautical and Aerospace Sciences FROM THE EARTH TO SPACE WITH NACA/NASA. M. Leroy Spearman. NASA- Langley Research Center, Hampton, VA 23681 & Heidi Owens, Auburn University, Auburn, AL 36849. Leonardo da Vinci envisioned man-flight in the 15th century and designed a practical airplane concept in 1490. Many other pioneers proposed various types of flying machines over the next 400 years but it was not until December 17, 1903 that the Wright Brothers, at Kitty Hawk, NC, were credited with achieving the first manned-powered flight. Over the next 100 years, several factors have influenced advances in aviation. The use of aircraft by European nations in World War I resulted in concern that the U.S. was lagging in aviation developments. This lead to an act of the U.S. -

Download Download
1999/2000. Proceedings of the Indiana Academy of Science 108/109:122-144 MAMMALS OF THE GRAND CALUMET RIVER REGION John O. Whitaker, Jr.: Department of Life Sciences, Indiana State University, Terre Haute. Indiana 47809 ABSTRACT. At least 37 of the 57 species of mammals occurring in Indiana are found in the Calumet region. These include species as follows: the opossum, 3 shrews, 1 mole, 4 bats, 1 rabbit, 7 squirrels, the beaver, 10 mice, 9 carnivores and the white-tailed deer. Keywords: Mammals, Indiana, Calumet, distribution The objectives of this paper are to describe pation when humans populate the land be- the pre-settlement and present-day mammal cause they are more feared (bear, wolf, moun- communities of the Grand Calumet River ba- tain lion) than smaller animals, or they are sin and to discuss how dredging operations hunted and trapped (deer, elk, bison, fisher, may affect these communities. A further ob- beaver) more than smaller mammals. Also, jective is to present some restoration options they usually need larger tracts of undisturbed that might be implemented during the dredg- habitat. Smaller mammals live alongside hu- ing operations to enhance the mammal popu- mans more easily because they are not hunted lations of the area. and they can use smaller patches of habitat. The extirpated species are discussed below. PRE-SETTLEMENT/EARLY SETTLEMENT MAMMAL COMMUNITY EXTIRPATED SPECIES Pre-settlement records of mammals of American porcupine (Erethizon dorsa- northwest Indiana are scant and consist main- tum).—The American porcupine was clearly ly of diary records of explorers such as Mar- present in pre-settlement times; skeletal re- quette and LaSalle, and of trading-post fur re- mains were found by Rand & Rand (1951). -

MF2975 Vole Control in Lawns and Landscapes
Vole Control in Lawns and Landscapes Voles (Microtus spp.) are small mammals that occur runways are usually throughout Kansas. Sometimes referred to as “meadow hidden beneath mice,” voles are compact rodents with short tails (about an a protective layer inch long), stocky bodies, big heads, and short legs. Their of grass or other eyes are small and ears partially hidden. They are usually dense ground cover. brown or gray, but coloration varies widely. Small holes lead to Three species of voles occur in the state: woodland underground runways vole (Microtus pinetorum), meadow vole (Microtus and nesting areas. pennsylvanicus), and prairie vole (Microtus ochrogaster) Gnaw marks about 1/8 (Figure 1). Voles are small, weighing 1 to 2 ounces as inch wide and 3/8 inch adults. Overall body length varies from 3 to 5½ inches for long are found on trees the woodland vole, to about 4½ to 7 inches for meadow or woody vegetation. and prairie voles. These three vole species differ in color and size. Usually it is not necessary to distinguish between the species to control the damage. Figure 2. Surface runway system of the prairie vole Population Dynamics Large population fluctuations are characteristic of voles. Population peaks occur about every three to four years. These are not regular cycles and do not usually involve spectacular population explosions. Occasionally, Figure 1. Prairie vole populations swell for about a year before declining. Several factors contribute to the potential for dramatic population growth. Voles play an important role in the food chain. They provide a major part of the diet for many predators, • Voles do not hibernate but remain active throughout the including coyotes, hawks, owls, foxes, and snakes. -

Management Plan for the Woodland Vole (Microtus Pinetorum) in Canada
Species at Risk Act Management Plan Series Management Plan for the Woodland Vole (Microtus pinetorum) in Canada Woodland Vole Tri-departmental Template Management Plan 2015 Recommended citation: Environment Canada. 2015. Management Plan for the Woodland Vole (Microtus pinetorum) in Canada. Species at Risk Act Management Plan Series. Canadian Wildlife Service, Ottawa. iv + 18 pp. For copies of the management plan, or for additional information on species at risk, including the Committee on the Status of Endangered Wildlife in Canada (COSEWIC) Status Reports, residence descriptions, action plans, and other related recovery documents, please visit the Species at Risk (SAR) Public Registry1. Cover illustration: © Philip Myers Également disponible en français sous le titre « Plan de gestion du campagnol sylvestre (Microtus pinetorum) au Canada » © Her Majesty the Queen in Right of Canada, represented by the Minister of the Environment, 2015. All rights reserved. ISBN 978-0-660-03060-9 Catalogue no. En3-5/62-2015E-PDF Content (excluding the illustrations) may be used without permission, with appropriate credit to the source. 1 http://www.registrelep-sararegistry.gc.ca Management Plan for the Woodland Vole 2015 Preface The federal, provincial, and territorial government signatories under the Accord for the Protection of Species at Risk (1996)2 agreed to establish complementary legislation and programs that provide for effective protection of species at risk throughout Canada. Under the Species at Risk Act (S.C. 2002, c.29) (SARA), the federal competent ministers are responsible for the preparation of management plans for listed species of special concern and are required to report on progress within five years after the publication of the final document on the SAR Public Registry. -
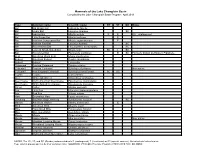
Mammals of the Lake Champlain Basin Compiled by the Lake Champlain Basin Program April 2013
Mammals of the Lake Champlain Basin Compiled by the Lake Champlain Basin Program April 2013 Type Common name Scientific name NY VT QC Notes Bat Big Brown Bat Eptesicus fuscus Bat Hoary Bat Lasiurus cinereus SC Bat Indiana Bat Myotis sodalis E E US - endangered Bat Little Brown Bat Myotis lucifugus E Bat Northern Long-eared Bat Myotis septentrionalis E Bat Eastern Red Bat Lasiurus borealis SC Bat Silver-haired Bat Lasionycteris noctivagans SC Bat Eastern Small-footed Bat Myotis leibii SC T SC Bat Tri-colored Bat Perimyotis subflavus E SC Formerly known as Eastern Pipistrelle Bear American Black Bear Ursus americanus Beaver American Beaver Castor canadensis Bobcat Bobcat Lynx rufus Chipmunk Eastern Chipmunk Tamias striatus Cottontail Eastern Cottontail Sylvilagus floridanus Non-native Cottontail New England Cottontail Sylvilagus transitionalis SC SC Coyote Coyote Canis latrans Deer White-tailed Deer Odocoileus virginianus Deermouse North American Deermouse Peromyscus maniculatus Deermouse White-footed Deermouse Peromyscus leucopus Fisher Fisher Martes pennanti Fox Gray Fox Urocyon cinereoargenteus Fox Red Fox Vulpes vulpes Hare Snowshoe Hare Lepus americanus Lemming Southern Bog Lemming Synaptomys cooperii SC Marten American Marten Martes americana E Mink American Mink Mustela vison Mole Hairy-tailed Mole Parascalops breweri Mole Star-nosed Mole Condylura cristata Moose Moose Alces americanus Mouse House Mouse Mus musculus Non-native Mouse Meadow Jumping Mouse Zapus hudsonius Mouse Woodland Jumping Mouse Napaeozapus insignis Muskrat Common Muskrat Ondata zibethicus Opossum Virginia Opossum Didelphis virginiana Otter North American River Otter Lutra canadensis NOTES: The NY, VT, and QC (Québec) columns indicate E (endangered), T (threatened) or SC (special concern). -

List of Native and Naturalized Fauna of Virginia
Virginia Department of Wildlife Resources List of Native and Naturalized Fauna of Virginia August, 2020 (* denotes naturalized species; ** denotes species native to some areas of Virginia and naturalized in other areas of Virginia) Common Name Scientific Name FISHES: Freshwater Fishes: Alabama Bass * Micropterus henshalli * Alewife Alosa pseudoharengus American Brook Lamprey Lampetra appendix American Eel Anguilla rostrata American Shad Alosa sapidissima Appalachia Darter Percina gymnocephala Ashy Darter Etheostoma cinereum Atlantic Sturgeon Acipenser oxyrhynchus Banded Darter Etheostoma zonale Banded Drum Larimus fasciatus Banded Killifish Fundulus diaphanus Banded Sculpin Cottus carolinae Banded Sunfish Ennaecanthus obesus Bigeye Chub Hybopsis amblops Bigeye Jumprock Moxostoma ariommum Bigmouth Chub Nocomis platyrhynchus Black Bullhead Ameiurus melas Black Crappie Pomoxis nigromaculatus Blacktip Jumprock Moxostoma cervinum Black Redhorse Moxostoma duquesnei Black Sculpin Cottus baileyi Blackbanded Sunfish Enneacanthus chaetodon Blacknose Dace Rhinichthys atratulus Blackside Dace Chrosomus cumberlandensis Blackside Darter Percina maculata Blotched Chub Erimystax insignis Blotchside Logperch Percina burtoni Blue Catfish * Ictalurus furcatus * Blue Ridge Sculpin Cottus caeruleomentum Blueback Herring Alosa aestivalis Bluebreast Darter Etheostoma camurum Bluegill Lepomis macrochirus Bluehead Chub Nocomis leptocephalus Blueside Darter Etheostoma jessiae Bluespar Darter Etheostoma meadiae Bluespotted Sunfish Enneacanthus gloriosus Bluestone -

Diet and Foraging Behaviors of Timber Rattlesnakes, Crotalus Horridus, in Eastern Virginia Author(S): Scott M
Diet and Foraging Behaviors of Timber Rattlesnakes, Crotalus horridus, in Eastern Virginia Author(s): Scott M. Goetz, Christopher E. Petersen, Robert K. Rose, John D. Kleopfer, and Alan H. Savitzky Source: Journal of Herpetology, 50(4):520-526. Published By: The Society for the Study of Amphibians and Reptiles DOI: http://dx.doi.org/10.1670/15-086 URL: http://www.bioone.org/doi/full/10.1670/15-086 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/terms_of_use. Usage of BioOne content is strictly limited to personal, educational, and non-commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Journal of Herpetology, Vol. 50, No. 4, 520–526, 2016 Copyright 2016 Society for the Study of Amphibians and Reptiles Diet and Foraging Behaviors of Timber Rattlesnakes, Crotalus horridus, in Eastern Virginia 1,2 3 1 4 1,5 SCOTT M. GOETZ, CHRISTOPHER E. PETERSEN, ROBERT K. ROSE, JOHN D. KLEOPFER, AND ALAN H. SAVITZKY 1Department of Biological Sciences, Old Dominion University, Norfolk, Virginia, USA 3Naval Facilities Engineering Command Atlantic, Norfolk, Virginia, USA 4Virginia Department of Game and Inland Fisheries, Charles City, Virginia, USA ABSTRACT.—During a 17-yr telemetry study, we examined the diet and ambush behavior of a population of Crotalus horridus in southeastern Virginia.