Acquired Resistance to Alectinib in ALK-Rearranged Lung Cancer Due
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Alectinib Provides a New Option for ALK-Positive NSCLC Patients After Progression on Crizotinib See Commentary on Page 239
Community Translations Edited by Jame Abraham, MD, FACP Alectinib provides a new option for ALK-positive NSCLC patients after progression on crizotinib See Commentary on page 239 n December 2015, alectinib became the third ALK inhibitor approved by the United States Food and What’s new, what’s important IDrug Administration for the treatment of non-small- The US Food and Drug Administration approved alectinib to cell lung cancer (NSCLC) that displays rearrangements of treat patients with metastatic ALK-positive non-small-cell lung the anaplastic lymphoma kinase (ALK) gene. Alectinib is cancer whose disease had progressed or who could not toler- a second-generation small molecule inhibitor of the ALK ate treatment with crizotinib. In clinical trials, alectinib showed a protein that joins ceritinib in providing a useful treatment partial response of 38%-44% and an average progression-free option for patients who have progressed on crizotinib, as survival of 11.2 months. In all, 61% of patients experienced a a result of its ability to target crizotinib-resistant mutant complete or partial reduction in their brain metastatic lesions, forms of the ALK protein. Alectinib also displays enhanced with a progression-free survival of of 9.1 months. The recom- penetrance of the blood-brain barrier, which improves ef- mended dose is 600 mg orally twice daily. cacy against central nervous system (CNS) metastases. The most common side effects are fatigue, constipation, swell- Te FDA awarded alectinib accelerated approval on the ing (edema), and muscle pain (myalgia). Treatment with alec- basis of 2 phase 2, single-arm clinical trials in patients tinib may cause sunburn when patients are exposed to sunlight, with ALK-positive NSCLC who had progressed on and pregnant women should be notifed of the possible risk to crizotinib therapy, a group of patients who have few avail- the fetus, and to use contraception throughout treatment and for able treatment options. -

Pharmaceutics of Oral Anticancer Agents and Stimulants
Pharmaceutics of oral anticancer agents and stimulants Maikel Herbrink The research in this thesis was performed at the Departments of Pharmacy & Pharma- cology and Medical Oncology & Clinical Pharmacology of the Netherlands Cancer Insti- tute – Antoni van Leeuwenhoek hospital, Amsterdam, the Netherlands. Printing of this thesis was financially supported by: Netherlands Cancer Institute Chipsoft BV Pfizer BV ISBN: 978-90-393-6997-5 Design, lay-out and print Gildeprint, Enschede © Maikel Herbrink, 2018 Pharmaceutics of oral anticancer agents and stimulants Farmaceutisch onderzoek naar oraal toegediende antikankermiddelen en stimulantia (met een samenvatting in het Nederlands) Proefschrift ter verkrijging van de graad van doctor aan de Universiteit Utrecht op gezag van de rector magnificus, prof. dr. H.R.B.M. Kummeling, ingevolge het besluit van het college voor promoties in het openbaar te verdedigen op woensdag 27 juni 2018 des middags te 4.15 uur door Maikel Herbrink geboren op 2 februari 1990 te Groningen Promotoren: Prof. dr. J.H. Beijnen Prof. dr. J.H.M. Schellens Copromotor: Dr. B. Nuijen Contents Preface 9 Chapter 1: Introduction 15 1.1 Variability in bioavailability of small molecular tyrosine 17 kinase inhibitors Cancer Treat Rev. 2015 May;41(5):412-422 1.2 Inherent formulation issues of kinase inhibitors 51 J Control Release. 2016 Oct;239:118-127 1.3 High-tech drugs in creaky formulations 81 Pharm Res. 2017 Sep;34(9):1751-1753 Chapter 2: Pharmaceutical analysis of oral anticancer drug substances 89 2.1. Thermal study of pazopanib hydrochloride 91 J Therm Anal Calorim. 2017 Dec;130(3):1491-1499 2.2. -

07052020 MR ASCO20 Curtain Raiser
Media Release New data at the ASCO20 Virtual Scientific Program reflects Roche’s commitment to accelerating progress in cancer care First clinical data from tiragolumab, Roche’s novel anti-TIGIT cancer immunotherapy, in combination with Tecentriq® (atezolizumab) in patients with PD-L1-positive metastatic non- small cell lung cancer (NSCLC) Updated overall survival data for Alecensa® (alectinib), in people living with anaplastic lymphoma kinase (ALK)-positive metastatic NSCLC Key highlights to be shared on Roche’s ASCO virtual newsroom, 29 May 2020, 08:00 CEST Basel, 7 May 2020 - Roche (SIX: RO, ROG; OTCQX: RHHBY) today announced that new data from clinical trials of 19 approved and investigational medicines across 21 cancer types, will be presented at the ASCO20 Virtual Scientific Program organised by the American Society of Clinical Oncology (ASCO), which will be held 29-31 May, 2020. A total of 120 abstracts that include a Roche medicine will be presented at this year's meeting. "At ASCO, we will present new data from many investigational and approved medicines across our broad oncology portfolio," said Levi Garraway, M.D., Ph.D., Roche's Chief Medical Officer and Head of Global Product Development. “These efforts exemplify our long-standing commitment to improving outcomes for people with cancer, even during these unprecedented times. By integrating our medicines and diagnostics together with advanced insights and novel platforms, Roche is uniquely positioned to deliver the healthcare solutions of the future." Together with its partners, Roche is pioneering a comprehensive approach to cancer care, combining new diagnostics and treatments with innovative, integrated data and access solutions for approved medicines that will both personalise and transform the outcomes of people affected by this deadly disease. -

Comparative Efficacy and Safety of Lorlatinib and Alectinib for ALK
cancers Systematic Review Comparative Efficacy and Safety of Lorlatinib and Alectinib for ALK-Rearrangement Positive Advanced Non-Small Cell Lung Cancer in Asian and Non-Asian Patients: A Systematic Review and Network Meta-Analysis Koichi Ando 1,2,* , Ryo Manabe 1, Yasunari Kishino 1, Sojiro Kusumoto 1, Toshimitsu Yamaoka 3 , Akihiko Tanaka 1, Tohru Ohmori 1,4 and Hironori Sagara 1 1 Division of Respiratory Medicine and Allergology, Department of Medicine, Showa University School of Medicine, 1-5-8 Hatanodai, Shinagawa-ku, Tokyo 142-8666, Japan; [email protected] (R.M.); [email protected] (Y.K.); [email protected] (S.K.); [email protected] (A.T.); [email protected] (T.O.); [email protected] (H.S.) 2 Division of Internal Medicine, Showa University Dental Hospital Medical Clinic, Senzoku Campus, Showa University, 2-1-1 Kita-senzoku, Ohta-ku, Tokyo 145-8515, Japan 3 Advanced Cancer Translational Research Institute, Showa University, 1-5-8 Hatanodai, Shinagawa-ku, Tokyo 142-8555, Japan; [email protected] 4 Department of Medicine, Division of Respiratory Medicine, Tokyo Metropolitan Health and Hospitals Corporation, Ebara Hospital, 4-5-10 Higashiyukigaya, Ohta-ku, Tokyo 145-0065, Japan * Correspondence: [email protected]; Tel.: +81-3-3784-8532 Citation: Ando, K.; Manabe, R.; Kishino, Y.; Kusumoto, S.; Yamaoka, Simple Summary: The treatment of anaplastic lymphoma kinase (ALK) rearrangement-positive T.; Tanaka, A.; Ohmori, T.; Sagara, H. (ALK-p) advanced non-small cell lung cancer (NSCLC) remains a challenge. -

Alecensa, INN-Alectinib
Package leaflet: Information for the patient Alecensa 150 mg hard capsules alectinib This medicine is subject to additional monitoring. This will allow quick identification of new safety information. You can help by reporting any side effects you may get. See the end of section 4 for how to report side effects. Read all of this leaflet carefully before you start taking this medicine - because it contains important information for you. ● Keep this leaflet. You may need to read it again. ● If you have any further questions, ask your doctor, pharmacist or nurse. ● This medicine has been prescribed for you only. Do not pass it on to others. It may harm them, even if their signs of illness are the same as yours. ● If you get any side effects, talk to your doctor, pharmacist or nurse. This includes any possible side effects not listed in this leaflet. See section 4. What is in this leaflet 1. What Alecensa is and what it is used for 2. What you need to know before you take Alecensa 3. How to take Alecensa 4. Possible side effects 5. How to store Alecensa 6. Contents of the pack and other information 1. What Alecensa is and what it is used for What Alecensa is Alecensa is a cancer medicine that contains the active substance alectinib. What Alecensa is used for Alecensa is used to treat adults with a type of lung cancer called ‘non-small cell lung cancer’ (‘NSCLC’). It is used if your lung cancer: ● is ‘ALK-positive’ - this means your cancer cells have a fault in a gene that makes an enzyme called ALK (‘anaplastic lymphoma kinase’), see ‘How Alecensa works’, below ● and is advanced. -

Alecensa® (Alectinib) Capsules(Genentech, Approved
tools Approved Drugs • Janssen Biotech, Inc. (janssenbiotech.com) cal will begin selling the once-a-day pill in announced that the FDA has granted the U.S. on Feb. 1, 2016. • The U.S. Food and Drug Administration accelerated approval to Darzalex™ (FDA) granted accelerated approval to (daratumumab injection) as a single • Millennium Pharmaceuticals (millennium. Alecensa® (alectinib) capsules (Genentech, agent for the treatment of patients with com) announced that the FDA has approved gene.com) for the treatment of patients with multiple myeloma who have received at Ninlaro® (ixazomib) in combination with anaplastic lymphoma kinase (ALK)- least three prior lines of therapy, including lenalidomide and dexamethasone for the positive metastatic non-small cell lung a proteasome inhibitor (PI) and an immuno- treatment of patients with multiple cancer (NSCLC) who have progressed on or are modulatory agent, or who are double- myeloma who have received at least one intolerant to crizotinib. Alecensa is an oral refractory to a PI and an immunomodula- prior therapy. Ixazomib is the first approved medication that blocks the activity of the ALK tory agent. oral proteasome inhibitor. protein, which may prevent NSCLC cells from growing and spreading. • The FDA approved Bristol-Myers Squibb’s • The FDA approved Bristol-Myers Squibb’s (bms.com) Empliciti™ (elotuzumab) in (bms.com) Opdivo® (nivolumab) Injection • Teva Pharmaceuticals Industries Ltd. combination with lenalidomide and for the treatment of advanced renal cell (tevausa.com) and Eagle Pharmaceuticals, Inc. dexamethasone for the treatment of carcinoma in patients who have received (eagleus.com) announced that the FDA has patients with multiple myeloma who have prior anti-angiogenic therapy. -

First-Line Treatment Options for Patients with Stage IV Non-Small Cell Lung Cancer with Driver Alterations
First-Line Treatment Options for Patients with Stage IV Non-Small Cell Lung Cancer with Driver Alterations Patients with stage IV non-small cell lung cancer Nonsquamous cell carcinoma and squamous cell carcinoma Activating EGFR mutation other Sensitizing (L858R/exon 19 MET exon 14 skipping than exon 20 insertion mutations, EGFR exon 20 mutation ALK rearrangement ROS1 rearrangement BRAF V600E mutation RET rearrangement NTRK rearrangement mutations KRAS alterations HER2 alterations NRG1 alterations deletion) EGFR mutation T790M, L858R or Ex19Del PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options Emerging target; no Emerging target; no Emerging target; no Platinum doublet † † † Osimertinib monotherapy S Afatinib monotherapy M M Alectinib S Entrectinib M Dabrafenib/trametinib M Capmatinib M Selpercatinib M Entrectinib M conclusions available conclusions available conclusions available chemotherapy ± bevacizumab Gefitinib with doublet Standard treatment based on Standard treatment based on Standard treatment based on M M M Brigatinib S Crizotinib M M Tepotinib M Pralsetinib* W Larotrectinib M chemotherapy non-driver mutation guideline non-driver mutation guideline non-driver mutation guideline If alectinib or brigatinib are not available If entrectinib or crizotinib are not available Standard treatment based on Standard treatment based on Standard treatment based on Dacomitinib monotherapy M Osimertinib W M M M Ceritinib S Ceritinib W non-driver mutation guideline non-driver mutation guideline non-driver mutation guideline Monotherapy with afatinib M Crizotinib S Lortlatinib W Standard treatment based on Erlotinib/ramucirumab M M non-driver mutation guideline Erlotinib/bevacizumab M Monotherapy with erlotinib M Strength of Recommendation Monotherapy with gefitinib M S Strong M Moderate W Weak Monotherapy with icotinib M Notes. -
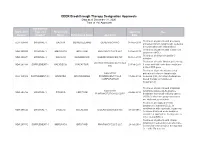
CDER Breakthrough Therapy Designation Approvals Data As of December 31, 2020 Total of 190 Approvals
CDER Breakthrough Therapy Designation Approvals Data as of December 31, 2020 Total of 190 Approvals Submission Application Type and Proprietary Approval Use Number Number Name Established Name Applicant Date Treatment of patients with previously BLA 125486 ORIGINAL-1 GAZYVA OBINUTUZUMAB GENENTECH INC 01-Nov-2013 untreated chronic lymphocytic leukemia in combination with chlorambucil Treatment of patients with mantle cell NDA 205552 ORIGINAL-1 IMBRUVICA IBRUTINIB PHARMACYCLICS LLC 13-Nov-2013 lymphoma (MCL) Treatment of chronic hepatitis C NDA 204671 ORIGINAL-1 SOVALDI SOFOSBUVIR GILEAD SCIENCES INC 06-Dec-2013 infection Treatment of cystic fibrosis patients age VERTEX PHARMACEUTICALS NDA 203188 SUPPLEMENT-4 KALYDECO IVACAFTOR 21-Feb-2014 6 years and older who have mutations INC in the CFTR gene Treatment of previously untreated NOVARTIS patients with chronic lymphocytic BLA 125326 SUPPLEMENT-60 ARZERRA OFATUMUMAB PHARMACEUTICALS 17-Apr-2014 leukemia (CLL) for whom fludarabine- CORPORATION based therapy is considered inappropriate Treatment of patients with anaplastic NOVARTIS lymphoma kinase (ALK)-positive NDA 205755 ORIGINAL-1 ZYKADIA CERITINIB 29-Apr-2014 PHARMACEUTICALS CORP metastatic non-small cell lung cancer (NSCLC) who have progressed on or are intolerant to crizotinib Treatment of relapsed chronic lymphocytic leukemia (CLL), in combination with rituximab, in patients NDA 206545 ORIGINAL-1 ZYDELIG IDELALISIB GILEAD SCIENCES INC 23-Jul-2014 for whom rituximab alone would be considered appropriate therapy due to other co-morbidities -

Newer-Generation EGFR Inhibitors in Lung Cancer: How Are They Best Used?
cancers Review Newer-Generation EGFR Inhibitors in Lung Cancer: How Are They Best Used? Tri Le 1 and David E. Gerber 1,2,3,* 1 Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, TX 75390-8852, USA; [email protected] 2 Department of Clinical Sciences, University of Texas Southwestern Medical Center, Dallas, TX 75390-8852, USA 3 Division of Hematology-Oncology, Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX 75390-8852, USA * Correspondence: [email protected]; Tel.: +1-214-648-4180; Fax: +1-214-648-1955 Received: 15 January 2019; Accepted: 4 March 2019; Published: 15 March 2019 Abstract: The FLAURA trial established osimertinib, a third-generation epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI), as a viable first-line therapy in non-small cell lung cancer (NSCLC) with sensitizing EGFR mutations, namely exon 19 deletion and L858R. In this phase 3 randomized, controlled, double-blind trial of treatment-naïve patients with EGFR mutant NSCLC, osimertinib was compared to standard-of-care EGFR TKIs (i.e., erlotinib or gefinitib) in the first-line setting. Osimertinib demonstrated improvement in median progression-free survival (18.9 months vs. 10.2 months; hazard ratio 0.46; 95% CI, 0.37 to 0.57; p < 0.001) and a more favorable toxicity profile due to its lower affinity for wild-type EGFR. Furthermore, similar to later-generation anaplastic lymphoma kinase (ALK) inhibitors, osimertinib has improved efficacy against brain metastases. Despite this impressive effect, the optimal sequencing of osimertinib, whether in the first line or as subsequent therapy after the failure of earlier-generation EGFR TKIs, is not clear. -

ALK Tyrosine Kinase Inhibitors, 5.01.538
PHARMACY POLICY – 5.01.538 ALK Tyrosine Kinase Inhibitors Effective Date: June 1, 2021 RELATED MEDICAL POLICIES: Last Revised: May 20, 2021 None Replaces: N/A Select a hyperlink below to be directed to that section. POLICY CRITERIA | CODING | RELATED INFORMATION EVIDENCE REVIEW | REFERENCES | HISTORY ∞ Clicking this icon returns you to the hyperlinks menu above. Introduction The anaplastic lymphoma kinase (ALK) gene provides instructions for making a specific kind of protein called ALK receptor tyrosine kinase. This protein helps cells communicate. When this gene is damaged, cell growth can get stuck in the “on” position and cells grow uncontrollably. Changes to the ALK gene can lead to non-small-cell lung cancer. Tyrosine kinase inhibitors block specific enzymes, essentially working to turn the cell growth to the “off” position. ALK tyrosine kinase inhibitors specifically targets cancers caused by changes to the ALK gene. This policy describes when this specific form of chemotherapy may be considered medically necessary. Note: The Introduction section is for your general knowledge and is not to be taken as policy coverage criteria. The rest of the policy uses specific words and concepts familiar to medical professionals. It is intended for providers. A provider can be a person, such as a doctor, nurse, psychologist, or dentist. A provider also can be a place where medical care is given, like a hospital, clinic, or lab. This policy informs them about when a service may be covered. Policy Coverage Criteria Drug Medical Necessity Alecensa® (alectinib) Alecensa® (alectinib) may be considered medically necessary for the treatment of adult patients with advanced or Drug Medical Necessity metastatic non-small cell lung cancer (NSCLC) that is anaplastic lymphoma kinase (ALK)-positive. -

Self Administered Oncology Agents Prior Authorization with Quantity
Self Administered Oncology Agents Prior Authorization with Quantity Limit Program Summary BCBSAL does not have the preferred product option. This program applies to Commercial, NetResults A series, SourceRx, and Health Insurance Marketplace formularies. OBJECTIVE The intent of the Self Administered Oncology Agents Prior Authorization (PA) program is to ensure appropriate selection of patients for treatment according to product labeling and/or clinical studies and/or clinical guidelines. The criteria considers appropriate indications as those supported in FDA approved labeling, National Comprehensive Cancer Network (NCCN) with level of evidence 1 or 2A recommendation, AHFS, or Drugdex with level of evidence of 1 or IIa. TARGET DRUGS Afinitor® (everolimus) Nerlynx™ (neratinib) Afinitor® Disperz (everolimus) Nexavar® (sorafenib) Alecensa® (alectinib) Ninlaro® (ixazomib) Alunbrig™ (brigatinib) Odomzo® (sonidegib) Bosulif® (bosutinib) Pomalyst® (pomalidomide) Cabometyx™ (cabozantinib) Revlimid® (lenalidomide) Calquence® (acalabrutinib) RubracaTM (rucaparib) Caprelsa® (vandetanib) Rydapt® (midostaurin) Cometriq™ (cabozantinib) Sprycel® (dasatinib) CotellicTM (cobimetinib) Stivarga® (regorafenib) Erivedge™ (vismodegib) Sutent® (sunitinib) Erleada™ (apalutamide) Sylatron® (peginterferon alfa-2b) Farydak® (panobinostat) Tafinlar® (dabrafenib) Gilotrif® (afatinib) TagrissoTM (osimertinib) Gleevec® (imatinib)a Tarceva® (erlotinib) Hexalen® (altretamine) Targretin® (bexarotene)a Hycamtin® (topotecan) Tasigna® (nilotinib) Ibrance® (palbociclib) Temodar® -

Sequential ALK Inhibitors Can Select for Lorlatinib-Resistant Compound ALK Mutations in ALK-Positive Lung Cancer
Published OnlineFirst April 12, 2018; DOI: 10.1158/2159-8290.CD-17-1256 RESEARCH ARTICLE Sequential ALK Inhibitors Can Select for Lorlatinib-Resistant Compound ALK Mutations in ALK-Positive Lung Cancer Satoshi Yoda1,2, Jessica J. Lin1,2, Michael S. Lawrence1,2,3, Benjamin J. Burke4, Luc Friboulet5, Adam Langenbucher1,2,3, Leila Dardaei1,2, Kylie Prutisto-Chang1, Ibiayi Dagogo-Jack1,2, Sergei Timofeevski4, Harper Hubbeling1,2, Justin F. Gainor1,2, Lorin A. Ferris1,2, Amanda K. Riley1, Krystina E. Kattermann1, Daria Timonina1, Rebecca S. Heist1,2, A. John Iafrate6, Cyril H. Benes1,2, Jochen K. Lennerz6, Mari Mino-Kenudson6, Jeffrey A. Engelman7, Ted W. Johnson4, Aaron N. Hata1,2, and Alice T. Shaw1,2 Downloaded from cancerdiscovery.aacrjournals.org on September 30, 2021. © 2018 American Association for Cancer Research. Published OnlineFirst April 12, 2018; DOI: 10.1158/2159-8290.CD-17-1256 ABSTRACT The cornerstone of treatment for advanced ALK-positive lung cancer is sequential therapy with increasingly potent and selective ALK inhibitors. The third-generation ALK inhibitor lorlatinib has demonstrated clinical activity in patients who failed previous ALK inhibi- tors. To define the spectrum ofALK mutations that confer lorlatinib resistance, we performed accel- erated mutagenesis screening of Ba/F3 cells expressing EML4–ALK. Under comparable conditions, N-ethyl-N-nitrosourea (ENU) mutagenesis generated numerous crizotinib-resistant but no lorlatinib- resistant clones harboring single ALK mutations. In similar screens with EML4–ALK containing single ALK resistance mutations, numerous lorlatinib-resistant clones emerged harboring compound ALK mutations. To determine the clinical relevance of these mutations, we analyzed repeat biopsies from lorlatinib-resistant patients.