Numerical Taxonomic Study of Some Tribes of Compositae (Subfamily Asteroideae) from Egypt
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
The Flora of Chad: a Checklist and Brief Analysis 1 Doi: 10.3897/Phytokeys.23.4752 Research Article Launched to Accelerate Biodiversity Research
A peer-reviewed open-access journal PhytoKeys 23: 1–17 (2013) The Flora of Chad: a checklist and brief analysis 1 doi: 10.3897/phytokeys.23.4752 RESEARCH ARTICLE www.phytokeys.com Launched to accelerate biodiversity research The Flora of Chad: a checklist and brief analysis Giuseppe Brundu1, Ignazio Camarda1 1 Department of Science for Nature and Environmental Resources (DIPNET), University of Sassari, Via Piandanna 4, 07100 Sassari, Italy Corresponding author: Giuseppe Brundu ([email protected]) Academic editor: Sandra Knapp | Received 23 January 2013 | Accepted 9 May 2013 | Published 13 May 2013 Citation: Brundu G, Camarda I (2013) The Flora of Chad: a checklist and brief analysis. PhytoKeys 23: 1–17. doi: 10.3897/phytokeys.23.4752 Abstract A checklist of the flora of Chad has been compiled by the authors, based on literature, on-line data-bases, herbarium collections and land surveys (1998-2011). It counts 2,460 records, i.e. 2,288 species (including 128 autonyms), 83 subspecies, 81 varieties, 8 forms, while all the previous available information reported 1,600 species. They belong to 151 Families, with 48.7% of the taxa belonging to the 6 largest families, i.e. Poaceae (14.6%), Fabaceae (13.6%), Cyperaceae (7.0%), Asteraceae (6.2 %), Malvaceae (3.9%) and Rubiaceae (3.4%). A total number or 2,173 species (88.3%) are native to Chad, including 55 (2.2%) endemic species, while 274 (11.0%) are alien to Chad, and 13 (0.5%) are considered cryptogenic, i.e. of uncertain status. It represents a considerable update on previous knowledge on the alien flora of Chad that counted for 131 taxa (5.3%). -

Safety Assessment of Helianthus Annuus (Sunflower)-Derived Ingredients As Used in Cosmetics
Safety Assessment of Helianthus annuus (Sunflower)-Derived Ingredients as Used in Cosmetics Status: Draft Tentative Report for Panel Review Release Date: March 7, 2016 Panel Meeting: March 31-April 1, 2016 The 2016 Cosmetic Ingredient Review Expert Panel members are: Chair, Wilma F. Bergfeld, M.D., F.A.C.P.; Donald V. Belsito, M.D.; Ronald A. Hill, Ph.D.; Curtis D. Klaassen, Ph.D.; Daniel C. Liebler, Ph.D.; James G. Marks, Jr., M.D.; Ronald C. Shank, Ph.D.; Thomas J. Slaga, Ph.D.; and Paul W. Snyder, D.V.M., Ph.D. The CIR Director is Lillian J. Gill, D.P.A. This report was prepared by Lillian C. Becker, Scientific Analyst/Writer. © Cosmetic Ingredient Review 1620 L Street, NW, Suite 1200 Washington, DC 20036-4702 ph 202.331.0651 fax 202.331.0088 [email protected] 1 Distributed for comment only -- do not cite or quote Commitment & Credibility since 1976 MEMORANDUM To: CIR Expert Panel and Liaisons From: Lillian C. Becker, M.S. Scientific Analyst and Writer Ivan J. Boyer, PhD, DABT Senior Toxicologist Date: March 7, 2016 Subject: Helianthus annuus (Sunflower)-Derived Ingredients as Used in Cosmetics Attached is the tentative report of 13 Helianthus annuus (sunflower)-derived ingredients as used in cosmetics. [Helian122015Rep] All of these ingredients are derived from parts of the Helianathus annuus (sunflower) plant. The sunflower seed oils (with the exception of Ozonized Sunflower Seed Oil) were reviewed in the vegetable- derived oils report and are not included here. In December, 2015, the Panel issued an Insufficient Data Announcement with these data needs: • HRIPT of Hydrogenated Sunflower Seed Extract at 1% or greater • Method of manufacture, including clarification of the source material (whole plant vs “bark”), of Helianthus Annuus (Sunflower) Extract • Composition of these ingredients, especially protein content (including 2S albumin) Impurities The Council submitted summaries of HRIPTs and use studies of products containing Helianthus annuus (sunflower)-derived ingredients. -

Seed Ecology Iii
SEED ECOLOGY III The Third International Society for Seed Science Meeting on Seeds and the Environment “Seeds and Change” Conference Proceedings June 20 to June 24, 2010 Salt Lake City, Utah, USA Editors: R. Pendleton, S. Meyer, B. Schultz Proceedings of the Seed Ecology III Conference Preface Extended abstracts included in this proceedings will be made available online. Enquiries and requests for hardcopies of this volume should be sent to: Dr. Rosemary Pendleton USFS Rocky Mountain Research Station Albuquerque Forestry Sciences Laboratory 333 Broadway SE Suite 115 Albuquerque, New Mexico, USA 87102-3497 The extended abstracts in this proceedings were edited for clarity. Seed Ecology III logo designed by Bitsy Schultz. i June 2010, Salt Lake City, Utah Proceedings of the Seed Ecology III Conference Table of Contents Germination Ecology of Dry Sandy Grassland Species along a pH-Gradient Simulated by Different Aluminium Concentrations.....................................................................................................................1 M Abedi, M Bartelheimer, Ralph Krall and Peter Poschlod Induction and Release of Secondary Dormancy under Field Conditions in Bromus tectorum.......................2 PS Allen, SE Meyer, and K Foote Seedling Production for Purposes of Biodiversity Restoration in the Brazilian Cerrado Region Can Be Greatly Enhanced by Seed Pretreatments Derived from Seed Technology......................................................4 S Anese, GCM Soares, ACB Matos, DAB Pinto, EAA da Silva, and HWM Hilhorst -

Barcoding the Asteraceae of Tennessee, Tribe Coreopsideae
Schilling, E.E., N. Mattson, and A. Floden. 2014. Barcoding the Asteraceae of Tennessee, tribe Coreopsideae. Phytoneuron 2014-101: 1–6. Published 20 October 2014. ISSN 2153 733X BARCODING THE ASTERACEAE OF TENNESSEE, TRIBE COREOPSIDEAE EDWARD E. SCHILLING, NICHOLAS MATTSON, AARON FLODEN Herbarium TENN Department of Ecology & Evolutionary Biology University of Tennessee Knoxville, Tennessee 37996 [email protected]; [email protected] ABSTRACT Results from barcoding studies of tribe Coreopsideae for the Tennessee flora using the nuclear ribosomal ITS marker are presented and include the first complete reports for 2 of the 20 species of the tribe that occur in the state, as well as updated reports for several others. Sequence data from the ITS region separate most of the species of Bidens in Tennessee from one another, but species of Coreopsis, especially those of sect. Coreopsis, have ITS sequences that are identical (or nearly so) to at least one congener. Comparisons of sequence data to GenBank records are complicated by apparent inaccuracies of older sequences as well as potentially misidentified samples. Broad survey of C. lanceolata from across its range showed little variability, but the ITS sequence of a morphologically distinct sample from a Florida limestone glade area was distinct in lacking a length polymorphism that was present in other samples. Tribe Coreopsideae is part of the Heliantheae alliance and earlier was often included in an expanded Heliantheae (Anderberg et al. 2007) in which it was usually treated as a subtribe (Crawford et al. 2009). The tribe shows a small burst of diversity in the southeastern USA involving Bidens and Coreopsis sect. -

Zeitschrift Für Naturforschung / C / 54 (1999)
602 Notes 16a,19-Diacetoxy-ewf-kaurane, a New Sephadex LH-20 to separate terpenoids from fla Natural Diterpene from the Exudate of vonoid aglcyones. The terpenoid portion (3.6 g) Ozothamnus scutellifolius (Asteraceae) was subjected to repeated column chromatogra phy on “flash” Si-gel using binary mixtures of Fco. Javier Arriaga-Giner3, Angel Rumberob increasing polarity (from H ex-AcO Et 10:1 v/v to and Eckhard Wollenweberc-* CH2Cl2-MeOH 20:1 v/v) furnishing 1 (19 mg), 2 a Tabacalera S. A., Centro I + D, c/Embajadores 51, E-28012 Madrid, Spain (72 mg), 3 (12 mg), 4 (25 mg), 5 (12 mg), 6 (23 mg) b Departamento de Quimica Orgänica, Universidad and 7 (9 mg). Autönoma de Madrid. Cantoblanco, Mass spectra were measured on a HP 5890 at 70 E-28049 Madrid, Spain eV via GC-MS. NMR spectra were recorded on a c Institut für Botanik der Technischen Universität, Schnittspahnstraße 3, D-64287 Darmstadt, Germany. Bruker AC-300 (300/75.4 MHz) in CDC13 or Fax: 06151/166878. CD3OD solutions. Multiplicities were assigned E-mail: [email protected] through DEPT experiments. * Author for correspondance and reprint requests 16a,19-Diacetoxy-e/ir-kaurane (1): White pow Z. Naturforsch. 54c, 602-604 (1999); der, mp. 112- 114oC. [a]D= -52.5 (c=0.55, CHC13). received April 1/April 29, 1999 Rf =0.44 (Hex/AcOEt 10:1). MS m/z (% rel. int.): Ozothamnus scutellifolius, Asteraceae, Leaf and Stem 390 (M+, -), 330 (M+ -AcOH, 13%), 315 (M+ - Exudate, Diterpenoids, Novel Kaurane Derivative AcOH-Me, 2), 270 (M+ -2AcOH, 4), 257 (13), 242 Leaf and stem exudate of Ozothamnus scutellifolius, (7), 221 (14), 161 (16), 119 (23), 106 (47), 94 (100), previously reported to contain 21 flavonoid aglycones, 81 (24), 67 (16), 55 (22) and 43 (73). -

Coreopsideae Daniel J
Chapter42 Coreopsideae Daniel J. Crawford, Mes! n Tadesse, Mark E. Mort, "ebecca T. Kimball and Christopher P. "andle HISTORICAL OVERVIEW AND PHYLOGENY In a cladistic analysis of morphological features of Heliantheae by Karis (1993), Coreopsidinae were reported Morphological data to be an ingroup within Heliantheae s.l. The group was A synthesis and analysis of the systematic information on represented in the analysis by Isostigma, Chrysanthellum, tribe Heliantheae was provided by Stuessy (1977a) with Cosmos, and Coreopsis. In a subsequent paper (Karis and indications of “three main evolutionary lines” within "yding 1994), the treatment of Coreopsidinae was the the tribe. He recognized ! fteen subtribes and, of these, same as the one provided above except for the follow- Coreopsidinae along with Fitchiinae, are considered ing: Diodontium, which was placed in synonymy with as constituting the third and smallest natural grouping Glossocardia by "obinson (1981), was reinstated following within the tribe. Coreopsidinae, including 31 genera, the work of Veldkamp and Kre# er (1991), who also rele- were divided into seven informal groups. Turner and gated Glossogyne and Guerreroia as synonyms of Glossocardia, Powell (1977), in the same work, proposed the new tribe but raised Glossogyne sect. Trionicinia to generic rank; Coreopsideae Turner & Powell but did not describe it. Eryngiophyllum was placed as a synonym of Chrysanthellum Their basis for the new tribe appears to be ! nding a suit- following the work of Turner (1988); Fitchia, which was able place for subtribe Jaumeinae. They suggested that the placed in Fitchiinae by "obinson (1981), was returned previously recognized genera of Jaumeinae ( Jaumea and to Coreopsidinae; Guardiola was left as an unassigned Venegasia) could be related to Coreopsidinae or to some Heliantheae; Guizotia and Staurochlamys were placed in members of Senecioneae. -

Studies Into the Genetic Architecture of C3-C4 Characteristics in Moricandia
Studies into the genetic architecture of C3-C4 characteristics in Moricandia Inaugural dissertation for the attainment of the title of doctor in the Faculty of Mathematics and Natural Sciences at the Heinrich Heine University Düsseldorf presented by Meng-Ying Lin from Taipei, Taiwan Düsseldorf, January 2020 From the institute of Plant Biochemistry at the Heinrich Heine University Düsseldorf Published by the permission of the Faculty of Mathematics and Natural Sciences at Heinrich Heine University Düsseldorf Supervisor: Prof. Dr. Andreas P.M. Weber Co-supervisor: Prof. Dr. Miltos Tsiantis Date of the oral examination: 05.05.2020 Declaration of the Doctoral Dissertation I herewith declare under oath that this dissertation was the result of my own work without any unauthorized help in compliance with the “Principles for the Safeguarding of Good Scientific Practice at Heinrich Heine University Düsseldorf”. This dissertation has never been submitted in this or similar format to any other institutions. I have not previously failed a doctoral examination procedure. Düsseldorf, 23.12.2019 Meng-Ying Lin Table of Contents I. Summary ............................................................................................................................... 1 II. Introduction ......................................................................................................................... 3 1. Motivation ...................................................................................................................................... -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -

17 Tribo Gnaphalieae (Cass.) Lecoq
17 Tribo Gnaphalieae (Cass.) Lecoq. & Juill. Leonardo Paz Deble SciELO Books / SciELO Livros / SciELO Libros DEBLE, L.P. Tribo Gnaphalieae (Cass.) Lecoq. & Juill. In: ROQUE, N. TELES, A.M., and NAKAJIMA, J.N., comp. A família Asteraceae no Brasil: classificação e diversidade [online]. Salvador: EDUFBA, 2017, pp. 131-137. ISBN: 978-85-232-1999-4. https://doi.org/10.7476/9788523219994.0019. All the contents of this work, except where otherwise noted, is licensed under a Creative Commons Attribution 4.0 International license. Todo o conteúdo deste trabalho, exceto quando houver ressalva, é publicado sob a licença Creative Commons Atribição 4.0. Todo el contenido de esta obra, excepto donde se indique lo contrario, está bajo licencia de la licencia Creative Commons Reconocimento 4.0. 17 TRIBO GNAPHALIEAE (CASS.) LECOQ. & JUILL. Leonardo Paz Deble A tribo Gnaphalieae é composta de 180-190 gêneros e cerca de 1.240 espécies, com distribuição quase cosmopolita, com maior diversidade na África do Sul, Ásia, Austrália e América do Sul (BAYER et al., 2007; DILLON; SAGASTEGUI, 1991; WARD et al., 2009). Para a América do Sul, Dillon e Sagastegui (1991) reconheceram 2 centros de diversidade: a Cordilheira dos Andes e o Sudeste do Brasil e regiões adjacentes. Tradicionalmente, Gnaphalieae foi subordinada à tribo Inuleae, sen- do reconhecida como um dos grupos de mais difícil identificação dentro da família, dadas a homogeneidade dos caracteres vegetativos e a escassa variação dos caracteres florais (CABRERA, 1961). Estudos recentes demons- traram que as Gnaphalieae encerram grupo monofilético posicionado na base da subfamília Asteroideae (ANDERBERG, 2009; ANDERBERG et al., 2005; FUNK et al., 2005). -

Chromosome Numbers in Compositae, XII: Heliantheae
SMITHSONIAN CONTRIBUTIONS TO BOTANY 0 NCTMBER 52 Chromosome Numbers in Compositae, XII: Heliantheae Harold Robinson, A. Michael Powell, Robert M. King, andJames F. Weedin SMITHSONIAN INSTITUTION PRESS City of Washington 1981 ABSTRACT Robinson, Harold, A. Michael Powell, Robert M. King, and James F. Weedin. Chromosome Numbers in Compositae, XII: Heliantheae. Smithsonian Contri- butions to Botany, number 52, 28 pages, 3 tables, 1981.-Chromosome reports are provided for 145 populations, including first reports for 33 species and three genera, Garcilassa, Riencourtia, and Helianthopsis. Chromosome numbers are arranged according to Robinson’s recently broadened concept of the Heliantheae, with citations for 212 of the ca. 265 genera and 32 of the 35 subtribes. Diverse elements, including the Ambrosieae, typical Heliantheae, most Helenieae, the Tegeteae, and genera such as Arnica from the Senecioneae, are seen to share a specialized cytological history involving polyploid ancestry. The authors disagree with one another regarding the point at which such polyploidy occurred and on whether subtribes lacking higher numbers, such as the Galinsoginae, share the polyploid ancestry. Numerous examples of aneuploid decrease, secondary polyploidy, and some secondary aneuploid decreases are cited. The Marshalliinae are considered remote from other subtribes and close to the Inuleae. Evidence from related tribes favors an ultimate base of X = 10 for the Heliantheae and at least the subfamily As teroideae. OFFICIALPUBLICATION DATE is handstamped in a limited number of initial copies and is recorded in the Institution’s annual report, Smithsonian Year. SERIESCOVER DESIGN: Leaf clearing from the katsura tree Cercidiphyllumjaponicum Siebold and Zuccarini. Library of Congress Cataloging in Publication Data Main entry under title: Chromosome numbers in Compositae, XII. -
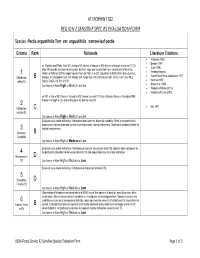
1 B 2 C 3 B 4 D 5 D
ATTACHMENT SS2 REGION 2 SENSITIVE SPECIES EVALUATION FORM Species: Pectis angustifolia Torr. var. angustifolia narrow-leaf pectis Criteria Rank Rationale Literature Citations • Anderson 1950 • Barkley 1991 se (Goshen and Platte Cos) WY, e into w NE; thence s through w KS; thence w through e and sw (?) CO. • Clark 1996 Most NE records are from the sw corner, but Keil maps one record from the n-central part of the state. 1 Weber & Wittman (2001b) report species from La Plata in w CO. [Vouchers at KANU from Baca, Lincoln, • Freeman in prep. • Great Plains Flora Association 1977 Distribution B Morgan, and Sedgwick Cos, CO; Meade and Trego Cos, KS; and Deuel, Keith, and Lincoln Cos, NE.] within R2 Status: G4G5; KS SH; WY S1 • Hartman 1997 • Shaw et al. 1989 Confidence in Rank High or Medium or Low • Weber & Wittman 2001a • Weber & Witman 2001b se WY, e into w NE; thence s through w KS; thence s w-most TX into n Mexico; thence n throughout NM; thence n through e CO, and w through n AZ and se-most UT. 2 • Keil 1977 Distribution C outside R2 Confidence in Rank High or Medium or Low Evaluator was unable to find any information about species’ dispersal capability. Seed is equipped with a pappus and may be dispersed by wind or perhaps water, during heavy rains. Dispersal is probably limited by 3 habitat requirements. Dispersal B Capability Confidence in Rank High or Medium or Low Evaluator was unable to find any information on species’ abundance within R2. Species does not appear to 4 be particularly abundant where encountered in CO and populations may face local extinction. -

Ndhf Sequence Evolution and the Major Clades in the Sunflower Family KI-JOONG KIM* and ROBERT K
Proc. Natl. Acad. Sci. USA Vol. 92, pp. 10379-10383, October 1995 Evolution ndhF sequence evolution and the major clades in the sunflower family KI-JOONG KIM* AND ROBERT K. JANSENt Department of Botany, University of Texas, Austin, TX 78713-7640 Communicated by Peter H. Raven, Missouri Botanical Garden, St. Louis, MO, June 21, 1995 ABSTRACT An extensive sequence comparison of the either too short or too conserved to provide adequate numbers chloroplast ndhF gene from all major clades of the largest of characters in recently evolved families. A number of alter- flowering plant family (Asteraceae) shows that this gene native genes have been suggested as potential candidates for provides -3 times more phylogenetic information than rbcL. phylogenetic comparisons at lower taxonomic levels (9). The This is because it is substantially longer and evolves twice as phylogenetic utility of one of these, matK, has been recently fast. The 5' region (1380 bp) ofndhF is very different from the demonstrated (10). Comparison of sequences of two chloro- 3' region (855 bp) and is similar to rbcL in both the rate and plast genomes (rice and tobacco), however, revealed only two the pattern of sequence change. The 3' region is more A+T- genes, rpoCl and ndhF, that are considerably longer and evolve rich, has higher levels of nonsynonymous base substitution, faster than rbcL (9, 11). We selected ndhF because it is longer and shows greater transversion bias at all codon positions. and evolves slightly faster than rpoCl (11), because rpoCl has These differences probably reflect different functional con- an intron that may require additional effort in DNA amplifi- straints on the 5' and 3' regions of nduhF.