Analysis of Gene Expression in Wild Type and Notch1 Mutant Retinal Cells by Single Cell Profiling Karolina Mizeracka1, Jeffrey M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Role of IL-1 in Adipose Browning and Muscle Wasting in CKD-Associated Cachexia
bioRxiv preprint doi: https://doi.org/10.1101/2021.02.05.429984; this version posted February 7, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The role of IL-1 in adipose browning and muscle wasting in CKD-associated cachexia Wai W Cheung1*, Ronghao Zheng2*, Sheng Hao3, Zhen Wang4, Alex Gonzalez1, Ping Zhou5, Hal M Hoffman6, Robert H Mak1 1 Pediatric Nephrology, Rady Children’s Hospital San Diego, University of California, San Diego, USA 2 Department of Pediatric Nephrology, Rheumatology, and Immunology, Maternal and Child Health Hospital of Hubei Province, TongJi Medical College, Huazhong University of Science and Technology, Wuhan, China 3Department of Nephrology and Rheumatology, Shanghai Children’s Hospital, Shanghai Jiao Tong University, Shanghai, China 4Department of Pediatrics, Shanghai General Hospital, Shanghai Jiao Tong University, Shanghai, China 5 Department of Pediatrics, the Second Affiliated Hospital of Harbin Medical University, Harbin, China 6 Department of Pediatrics, University of California, San Diego, USA * These authors contributed equally to this work. Correspondence: Robert H Mak Division of Pediatric Nephrology Rady Children’s Hospital University of California, San Diego 9500 Gilman Drive, MC0831, La Jolla, California 92093-0831, USA P: 858-822-6717 F: 858-822-6776 E-mail: [email protected] Running title: Anakinra attenuates uremic cachexia bioRxiv preprint doi: https://doi.org/10.1101/2021.02.05.429984; this version posted February 7, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. -

A Prelude to the Proximity Interaction Mapping of CXXC5
www.nature.com/scientificreports OPEN A prelude to the proximity interaction mapping of CXXC5 Gamze Ayaz1,4,6*, Gizem Turan1,6, Çağla Ece Olgun1,6, Gizem Kars1, Burcu Karakaya1, Kerim Yavuz1, Öykü Deniz Demiralay1, Tolga Can2, Mesut Muyan1,3* & Pelin Yaşar1,5,6 CXXC5 is a member of the zinc-fnger CXXC family proteins that interact with unmodifed CpG dinucleotides through a conserved ZF-CXXC domain. CXXC5 is involved in the modulation of gene expressions that lead to alterations in diverse cellular events. However, the underlying mechanism of CXXC5-modulated gene expressions remains unclear. Proteins perform their functions in a network of proteins whose identities and amounts change spatiotemporally in response to various stimuli in a lineage-specifc manner. Since CXXC5 lacks an intrinsic transcription regulatory function or enzymatic activity but is a DNA binder, CXXC5 by interacting with proteins could act as a scafold to establish a chromatin state restrictive or permissive for transcription. To initially address this, we utilized the proximity-dependent biotinylation approach. Proximity interaction partners of CXXC5 include DNA and chromatin modifers, transcription factors/co-regulators, and RNA processors. Of these, CXXC5 through its CXXC domain interacted with EMD, MAZ, and MeCP2. Furthermore, an interplay between CXXC5 and MeCP2 was critical for a subset of CXXC5 target gene expressions. It appears that CXXC5 may act as a nucleation factor in modulating gene expressions. Providing a prelude for CXXC5 actions, our results could also contribute to a better understanding of CXXC5-mediated cellular processes in physiology and pathophysiology. Te methylation of mammalian genomic DNA, which predominantly arises post-replicatively at the 5’ posi- tion of the cytosine base in the context of CpG dinucleotides, varies across cell types, developmental stages, physiological and pathophysiological conditions 1. -

THE ROLE of HOMEODOMAIN TRANSCRIPTION FACTOR IRX5 in CARDIAC CONTRACTILITY and HYPERTROPHIC RESPONSE by © COPYRIGHT by KYOUNG H
THE ROLE OF HOMEODOMAIN TRANSCRIPTION FACTOR IRX5 IN CARDIAC CONTRACTILITY AND HYPERTROPHIC RESPONSE By KYOUNG HAN KIM A THESIS SUBMITTED IN CONFORMITY WITH THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY GRADUATE DEPARTMENT OF PHYSIOLOGY UNIVERSITY OF TORONTO © COPYRIGHT BY KYOUNG HAN KIM (2011) THE ROLE OF HOMEODOMAIN TRANSCRIPTION FACTOR IRX5 IN CARDIAC CONTRACTILITY AND HYPERTROPHIC RESPONSE KYOUNG HAN KIM DOCTOR OF PHILOSOPHY GRADUATE DEPARTMENT OF PHYSIOLOGY UNIVERSITY OF TORONTO 2011 ABSTRACT Irx5 is a homeodomain transcription factor that negatively regulates cardiac fast transient + outward K currents (Ito,f) via the KV4.2 gene and is thereby a major determinant of the transmural repolarization gradient. While Ito,f is invariably reduced in heart disease and changes in Ito,f can modulate both cardiac contractility and hypertrophy, less is known about a functional role of Irx5, and its relationship with Ito,f, in the normal and diseased heart. Here I show that Irx5 plays crucial roles in the regulation of cardiac contractility and proper adaptive hypertrophy. Specifically, Irx5-deficient (Irx5-/-) hearts had reduced cardiac contractility and lacked the normal regional difference in excitation-contraction with decreased action potential duration, Ca2+ transients and myocyte shortening in sub-endocardial, but not sub-epicardial, myocytes. In addition, Irx5-/- mice showed less cardiac hypertrophy, but increased interstitial fibrosis and greater contractility impairment following pressure overload. A defect in hypertrophic responses in Irx5-/- myocardium was confirmed in cultured neonatal mouse ventricular myocytes, exposed to norepinephrine while being restored with Irx5 replacement. Interestingly, studies using mice ii -/- virtually lacking Ito,f (i.e. KV4.2-deficient) showed that reduced contractility in Irx5 mice was completely restored by loss of KV4.2, whereas hypertrophic responses to pressure-overload in hearts remained impaired when both Irx5 and Ito,f were absent. -
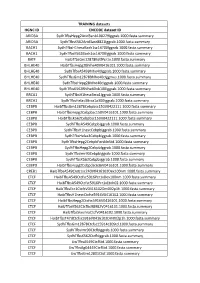
TRAINING Datasets HGNC ID ENCODE Dataset ID ARID3A
TRAINING datasets HGNC ID ENCODE dataset ID ARID3A SydhT+sHepg2Arid3anb100279Iggrab.1000.fasta.summary ARID3A SydhT+sK562Arid3asC8821Iggrab.1000.fasta.summary BACH1 SydhT+sH1hesCBaCh1sC14700Iggrab.1000.fasta.summary BACH1 SydhT+sK562BaCh1sC14700Iggrab.1000.fasta.summary BATF HaibT+sGm12878BaJPCr1x.1000.fasta.summary BHLHE40 HaibT+sHepg2Bhlhe40V0416101.1000.fasta.summary BHLHE40 SydhT+sA549Bhlhe40Iggrab.1000.fasta.summary BHLHE40 SydhT+sGm12878Bhlhe40CIggmus.1000.fasta.summary BHLHE40 SydhT+sHepg2Bhlhe40CIggrab.1000.fasta.summary BHLHE40 SydhT+sK562Bhlhe40nb100Iggrab.1000.fasta.summary BRCA1 SydhT+sH1hesCBrCa1Iggrab.1000.fasta.summary BRCA1 SydhT+sHelas3BrCa1a300Iggrab.1000.fasta.summary CEBPB HaibT+sGm12878CebpbsC150V0422111.1000.fasta.summary CEBPB HaibT+sHepg2CebpbsC150V0416101.1000.fasta.summary CEBPB HaibT+sK562CebpbsC150V0422111.1000.fasta.summary CEBPB SydhT+sA549CebpbIggrab.1000.fasta.summary CEBPB SydhT+sH1hesCCebpbIggrab.1000.fasta.summary CEBPB SydhT+sHelas3CebpbIggrab.1000.fasta.summary CEBPB SydhT+sHepg2CebpbForsklnStd.1000.fasta.summary CEBPB SydhT+sHepg2CebpbIggrab.1000.fasta.summary CEBPB SydhT+sImr90CebpbIggrab.1000.fasta.summary CEBPB SydhT+sK562CebpbIggrab.1000.fasta.summary CEBPD HaibT+sHepg2CebpdsC636V0416101.1000.fasta.summary CREB1 HaibT+sA549Creb1sC240V0416102Dex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xDex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xEtoh02.1000.fasta.summary CTCF HaibT+sECC1CtCfCV0416102Dm002p1h.1000.fasta.summary CTCF HaibT+sH1hesCCtCfsC5916V0416102.1000.fasta.summary -

Sanjay Kumar Gupta
The human CCHC-type Zinc Finger Nucleic Acid Binding Protein (CNBP) binds to the G-rich elements in target mRNA coding sequences and promotes translation Das humane CCHC-Typ-Zinkfinger-Nukleinsäure-Binde-Protein (CNBP) bindet an G-reiche Elemente in der kodierenden Sequenz seiner Ziel-mRNAs und fördert deren Translation Doctoral thesis for a doctoral degree at the Graduate School of Life Sciences, Julius-Maximilians-Universität WürzBurg, Section: Biomedicine suBmitted By Sanjay Kumar Gupta from Varanasi, India WürzBurg, 2016 1 Submitted on: …………………………………………………………..…….. Office stamp Members of the Promotionskomitee: Chairperson: Prof. Dr. Alexander Buchberger Primary Supervisor: Dr. Stefan Juranek Supervisor (Second): Prof. Dr. Utz Fischer Supervisor (Third): Dr. Markus Landthaler Date of Public Defence: …………………………………………….………… Date of Receipt of Certificates: ………………………………………………. 2 Summary The genetic information encoded with in the genes are transcribed and translated to give rise to the functional proteins, which are building block of a cell. At first, it was thought that the regulation of gene expression particularly occurs at the level of transcription By various transcription factors. Recent discoveries have shown the vital role of gene regulation at the level of RNA also known as post-transcriptional gene regulation (PTGR). Apart from non-coding RNAs e.g. micro RNAs, various RNA Binding proteins (RBPs) play essential role in PTGR. RBPs have been implicated in different stages of mRNA life cycle ranging from splicing, processing, transport, localization and decay. In last 20 years studies have shown the presence of hundreds of RBPs across eukaryotic systems many of which are widely conserved. Given the rising numBer of RBPs and their link to human diseases it is quite evident that RBPs have major role in cellular processes and their regulation. -

Multifactorial Erβ and NOTCH1 Control of Squamous Differentiation and Cancer
Multifactorial ERβ and NOTCH1 control of squamous differentiation and cancer Yang Sui Brooks, … , Karine Lefort, G. Paolo Dotto J Clin Invest. 2014;124(5):2260-2276. https://doi.org/10.1172/JCI72718. Research Article Oncology Downmodulation or loss-of-function mutations of the gene encoding NOTCH1 are associated with dysfunctional squamous cell differentiation and development of squamous cell carcinoma (SCC) in skin and internal organs. While NOTCH1 receptor activation has been well characterized, little is known about how NOTCH1 gene transcription is regulated. Using bioinformatics and functional screening approaches, we identified several regulators of the NOTCH1 gene in keratinocytes, with the transcription factors DLX5 and EGR3 and estrogen receptor β (ERβ) directly controlling its expression in differentiation. DLX5 and ERG3 are required for RNA polymerase II (PolII) recruitment to the NOTCH1 locus, while ERβ controls NOTCH1 transcription through RNA PolII pause release. Expression of several identified NOTCH1 regulators, including ERβ, is frequently compromised in skin, head and neck, and lung SCCs and SCC-derived cell lines. Furthermore, a keratinocyte ERβ–dependent program of gene expression is subverted in SCCs from various body sites, and there are consistent differences in mutation and gene-expression signatures of head and neck and lung SCCs in female versus male patients. Experimentally increased ERβ expression or treatment with ERβ agonists inhibited proliferation of SCC cells and promoted NOTCH1 expression and squamous differentiation both in vitro and in mouse xenotransplants. Our data identify a link between transcriptional control of NOTCH1 expression and the estrogen response in keratinocytes, with implications for differentiation therapy of squamous cancer. Find the latest version: https://jci.me/72718/pdf Research article Multifactorial ERβ and NOTCH1 control of squamous differentiation and cancer Yang Sui Brooks,1,2 Paola Ostano,3 Seung-Hee Jo,1,2 Jun Dai,1,2 Spiro Getsios,4 Piotr Dziunycz,5 Günther F.L. -

Gene Therapy in Cancer Treatment: Why Go Nano?
pharmaceutics Review Gene Therapy in Cancer Treatment: Why Go Nano? Catarina Roma-Rodrigues 1 , Lorenzo Rivas-García 1,2 , Pedro V. Baptista 1,* and Alexandra R. Fernandes 1,* 1 UCIBIO, Departamento de Ciências da Vida, Faculdade de Ciências e Tecnologia, Campus de Caparica, 2829-516 Caparica, Portugal; [email protected] (C.R.-R.); [email protected] (L.R.-G.) 2 Biomedical Research Centre, Institute of Nutrition and Food Technology, Department of Physiology, Faculty of Pharmacy, University of Granada, Avda. del Conocimiento s/n. 18071 Armilla, Granada, Spain * Correspondence: [email protected] (P.V.B.); [email protected] (A.R.F.); Tel.: +351-212-948-530 (P.V.B. & A.R.F.) Received: 29 January 2020; Accepted: 3 March 2020; Published: 5 March 2020 Abstract: The proposal of gene therapy to tackle cancer development has been instrumental for the development of novel approaches and strategies to fight this disease, but the efficacy of the proposed strategies has still fallen short of delivering the full potential of gene therapy in the clinic. Despite the plethora of gene modulation approaches, e.g., gene silencing, antisense therapy, RNA interference, gene and genome editing, finding a way to efficiently deliver these effectors to the desired cell and tissue has been a challenge. Nanomedicine has put forward several innovative platforms to overcome this obstacle. Most of these platforms rely on the application of nanoscale structures, with particular focus on nanoparticles. Herein, we review the current trends on the use of nanoparticles designed for cancer gene therapy, including inorganic, organic, or biological (e.g., exosomes) variants, in clinical development and their progress towards clinical applications. -

The Zinc-Finger Protein CNBP Is Required for Forebrain Formation In
Development 130, 1367-1379 1367 © 2003 The Company of Biologists Ltd doi:10.1242/dev.00349 The zinc-finger protein CNBP is required for forebrain formation in the mouse Wei Chen1,2, Yuqiong Liang1, Wenjie Deng1, Ken Shimizu1, Amir M. Ashique1,2, En Li3 and Yi-Ping Li1,2,* 1Department of Cytokine Biology, The Forsyth Institute, Boston, MA 02115, USA 2Harvard-Forsyth Department of Oral Biology, Harvard School of Dental Medicine, Boston, MA 02115, USA 3Cardiovascular Research Center, Massachusetts General Hospital, Department of Medicine, Harvard Medical School, Charlestown, MA 02129, USA *Author for correspondence (e-mail: [email protected]) Accepted 19 December 2002 SUMMARY Mouse mutants have allowed us to gain significant insight (AME), headfolds and forebrain. In Cnbp–/– embryos, the into axis development. However, much remains to be visceral endoderm remains in the distal tip of the conceptus learned about the cellular and molecular basis of early and the ADE fails to form, whereas the node and notochord forebrain patterning. We describe a lethal mutation mouse form normally. A substantial reduction in cell proliferation strain generated using promoter-trap mutagenesis. The was observed in the anterior regions of Cnbp–/– embryos at mutants exhibit severe forebrain truncation in homozygous gastrulation and neural-fold stages. In these regions, Myc mouse embryos and various craniofacial defects in expression was absent, indicating CNBP targets Myc in heterozygotes. We show that the defects are caused by rostral head formation. Our findings demonstrate that disruption of the gene encoding cellular nucleic acid Cnbp is essential for the forebrain induction and binding protein (CNBP); Cnbp transgenic mice were able specification. -

Supplementary Materials
Supplementary Materials: Supplemental Table 1 Abbreviations FMDV Foot and Mouth Disease Virus FMD Foot and Mouth Disease NC Non-treated Control DEGs Differentially Expressed Genes RNA-seq High-throughput Sequencing of Mrna RT-qPCR Quantitative Real-time Reverse Transcriptase PCR TCID50 50% Tissue Culture Infective Doses CPE Cytopathic Effect MOI Multiplicity of Infection DMEM Dulbecco's Modified Eagle Medium FBS Fetal Bovine Serum PBS Phosphate Buffer Saline QC Quality Control FPKM Fragments per Kilo bases per Million fragments method GO Gene Ontology KEGG Kyoto Encyclopedia of Genes and Genomes R Pearson Correlation Coefficient NFKBIA NF-kappa-B Inhibitor alpha IL6 Interleukin 6 CCL4 C-C motif Chemokine 4 CXCL2 C-X-C motif Chemokine 2 TNF Tumor Necrosis Factor VEGFA Vascular Endothelial Growth Gactor A CCL20 C-C motif Chemokine 20 CSF2 Macrophage Colony-Stimulating Factor 2 GADD45B Growth Arrest and DNA Damage Inducible 45 beta MYC Myc proto-oncogene protein FOS Proto-oncogene c-Fos MCL1 Induced myeloid leukemia cell differentiation protein Mcl-1 MAP3K14 Mitogen-activated protein kinase kinase kinase 14 IRF1 Interferon regulatory factor 1 CCL5 C-C motif chemokine 5 ZBTB3 Zinc finger and BTB domain containing 3 OTX1 Orthodenticle homeobox 1 TXNIP Thioredoxin-interacting protein ZNF180 Znc Finger Protein 180 ZNF36 Znc Finger Protein 36 ZNF182 Zinc finger protein 182 GINS3 GINS complex subunit 3 KLF15 Kruppel-like factor 15 Supplemental Table 2 Primers for Verification of RNA-seq-detected DEGs with RT-qPCR TNF F: CGACTCAGTGCCGAGATCAA R: -

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

Open Dogan Phdthesis Final.Pdf
The Pennsylvania State University The Graduate School Eberly College of Science ELUCIDATING BIOLOGICAL FUNCTION OF GENOMIC DNA WITH ROBUST SIGNALS OF BIOCHEMICAL ACTIVITY: INTEGRATIVE GENOME-WIDE STUDIES OF ENHANCERS A Dissertation in Biochemistry, Microbiology and Molecular Biology by Nergiz Dogan © 2014 Nergiz Dogan Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy August 2014 ii The dissertation of Nergiz Dogan was reviewed and approved* by the following: Ross C. Hardison T. Ming Chu Professor of Biochemistry and Molecular Biology Dissertation Advisor Chair of Committee David S. Gilmour Professor of Molecular and Cell Biology Anton Nekrutenko Professor of Biochemistry and Molecular Biology Robert F. Paulson Professor of Veterinary and Biomedical Sciences Philip Reno Assistant Professor of Antropology Scott B. Selleck Professor and Head of the Department of Biochemistry and Molecular Biology *Signatures are on file in the Graduate School iii ABSTRACT Genome-wide measurements of epigenetic features such as histone modifications, occupancy by transcription factors and coactivators provide the opportunity to understand more globally how genes are regulated. While much effort is being put into integrating the marks from various combinations of features, the contribution of each feature to accuracy of enhancer prediction is not known. We began with predictions of 4,915 candidate erythroid enhancers based on genomic occupancy by TAL1, a key hematopoietic transcription factor that is strongly associated with gene induction in erythroid cells. Seventy of these DNA segments occupied by TAL1 (TAL1 OSs) were tested by transient transfections of cultured hematopoietic cells, and 56% of these were active as enhancers. Sixty-six TAL1 OSs were evaluated in transgenic mouse embryos, and 65% of these were active enhancers in various tissues. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated.