Analysis of the Genetic Diversity of the Coastal and Island Endangered Plant Species Elaeagnus Macrophylla Via Conserved DNA-Derived Polymorphism Marker
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

TIER2 SITE NAME ADDRESS PROCESS M Ns Garments Printing & Embroidery
TIER 2 MANUFACTURING SITES - Produced July 2021 TIER2 SITE NAME ADDRESS PROCESS Bangladesh Mns Garments Printing & Embroidery (Unit 2) House 305 Road 34 Hazirpukur Choydana National University Gazipur Manufacturer/Processor (A&E) American & Efird (Bd) Ltd Plot 659 & 660 93 Islampur Gazipur Manufacturer/Processor A G Dresses Ltd Ag Tower Plot 09 Block C Tongi Industrial Area Himardighi Gazipur Next Branded Component Abanti Colour Tex Ltd Plot S A 646 Shashongaon Enayetnagar Fatullah Narayanganj Manufacturer/Processor Aboni Knitwear Ltd Plot 169 171 Tetulzhora Hemayetpur Savar Dhaka 1340 Manufacturer/Processor Afrah Washing Industries Ltd Maizpara Taxi Track Area Pan - 4 Patenga Chottogram Manufacturer/Processor AKM Knit Wear Limited Holding No 14 Gedda Cornopara Ulail Savar Dhaka Next Branded Component Aleya Embroidery & Aleya Design Hose 40 Plot 808 Iqbal Bhaban Dhour Nishat Nagar Turag Dhaka 1230 Manufacturer/Processor Alim Knit (Bd) Ltd Nayapara Kashimpur Gazipur 1750 Manufacturer/Processor Aman Fashions & Designs Ltd Nalam Mirzanagar Asulia Savar Manufacturer/Processor Aman Graphics & Design Ltd Nazimnagar Hemayetpur Savar Dhaka Manufacturer/Processor Aman Sweaters Ltd Rajaghat Road Rajfulbaria Savar Dhaka Manufacturer/Processor Aman Winter Wears Ltd Singair Road Hemayetpur Savar Dhaka Manufacturer/Processor Amann Bd Plot No Rs 2497-98 Tapirbari Tengra Mawna Shreepur Gazipur Next Branded Component Amantex Limited Boiragirchala Sreepur Gazipur Manufacturer/Processor Ananta Apparels Ltd - Adamjee Epz Plot 246 - 249 Adamjee Epz Narayanganj -

Number 3, Spring 1998 Director’S Letter
Planning and planting for a better world Friends of the JC Raulston Arboretum Newsletter Number 3, Spring 1998 Director’s Letter Spring greetings from the JC Raulston Arboretum! This garden- ing season is in full swing, and the Arboretum is the place to be. Emergence is the word! Flowers and foliage are emerging every- where. We had a magnificent late winter and early spring. The Cornus mas ‘Spring Glow’ located in the paradise garden was exquisite this year. The bright yellow flowers are bright and persistent, and the Students from a Wake Tech Community College Photography Class find exfoliating bark and attractive habit plenty to photograph on a February day in the Arboretum. make it a winner. It’s no wonder that JC was so excited about this done soon. Make sure you check of themselves than is expected to seedling selection from the field out many of the special gardens in keep things moving forward. I, for nursery. We are looking to propa- the Arboretum. Our volunteer one, am thankful for each and every gate numerous plants this spring in curators are busy planting and one of them. hopes of getting it into the trade. preparing those gardens for The magnolias were looking another season. Many thanks to all Lastly, when you visit the garden I fantastic until we had three days in our volunteers who work so very would challenge you to find the a row of temperatures in the low hard in the garden. It shows! Euscaphis japonicus. We had a twenties. There was plenty of Another reminder — from April to beautiful seven-foot specimen tree damage to open flowers, but the October, on Sunday’s at 2:00 p.m. -

Agroforestry News Index Vol 1 to Vol 22 No 2
Agroforestry News Index Vol 1 to Vol 22 No 2 2 A.R.T. nursery ..... Vol 2, No 4, page 2 Acorns, edible from oaks ..... Vol 5, No 4, page 3 Aaron, J R & Richards: British woodland produce (book review) ..... Acorns, harvesting ..... Vol 5, No 4, Vol 1, No 4, page 34 page 3 Abies balsamea ..... Vol 8, No 2, page Acorns, nutritional composition ..... 31 Vol 5, No 4, page 4 Abies sibirica ..... Vol 8, No 2, page 31 Acorns, removing tannins from ..... Vol 5, No 4, page 4 Abies species ..... Vol 19, No 1, page 13 Acorns, shelling ..... Vol 5, No 4, page 3 Acca sellowiana ..... Vol 9, No 3, page 4 Acorns, utilisation ..... Vol 5, No 4, page 4 Acer macrophyllum ..... Vol 16, No 2, page 6 Acorus calamus ..... Vol 8, No 4, page 6 Acer pseudoplatanus ..... Vol 3, No 1, page 3 Actinidia arguta ..... Vol 1, No 4, page 10 Acer saccharum ..... Vol 16, No 1, page 3 Actinidia arguta, cultivars ..... Vol 1, No 4, page 14 Acer saccharum - strawberry agroforestry system ..... Vol 8, No 1, Actinidia arguta, description ..... Vol page 2 1, No 4, page 10 Acer species, with edible saps ..... Vol Actinidia arguta, drawings ..... Vol 1, 2, No 3, page 26 No 4, page 15 Achillea millefolium ..... Vol 8, No 4, Actinidia arguta, feeding & irrigaton page 5 ..... Vol 1, No 4, page 11 3 Actinidia arguta, fruiting ..... Vol 1, Actinidia spp ..... Vol 5, No 1, page 18 No 4, page 13 Actinorhizal plants ..... Vol 3, No 3, Actinidia arguta, nurseries page 30 supplying ..... Vol 1, No 4, page 16 Acworth, J M: The potential for farm Actinidia arguta, pests and diseases forestry, agroforestry and novel tree .... -
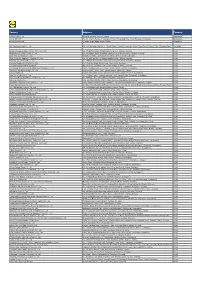
Factory Address Country
Factory Address Country Durable Plastic Ltd. Mulgaon, Kaligonj, Gazipur, Dhaka Bangladesh Lhotse (BD) Ltd. Plot No. 60&61, Sector -3, Karnaphuli Export Processing Zone, North Potenga, Chittagong Bangladesh Bengal Plastics Ltd. Yearpur, Zirabo Bazar, Savar, Dhaka Bangladesh ASF Sporting Goods Co., Ltd. Km 38.5, National Road No. 3, Thlork Village, Chonrok Commune, Korng Pisey District, Konrrg Pisey, Kampong Speu Cambodia Ningbo Zhongyuan Alljoy Fishing Tackle Co., Ltd. No. 416 Binhai Road, Hangzhou Bay New Zone, Ningbo, Zhejiang China Ningbo Energy Power Tools Co., Ltd. No. 50 Dongbei Road, Dongqiao Industrial Zone, Haishu District, Ningbo, Zhejiang China Junhe Pumps Holding Co., Ltd. Wanzhong Villiage, Jishigang Town, Haishu District, Ningbo, Zhejiang China Skybest Electric Appliance (Suzhou) Co., Ltd. No. 18 Hua Hong Street, Suzhou Industrial Park, Suzhou, Jiangsu China Zhejiang Safun Industrial Co., Ltd. No. 7 Mingyuannan Road, Economic Development Zone, Yongkang, Zhejiang China Zhejiang Dingxin Arts&Crafts Co., Ltd. No. 21 Linxian Road, Baishuiyang Town, Linhai, Zhejiang China Zhejiang Natural Outdoor Goods Inc. Xiacao Village, Pingqiao Town, Tiantai County, Taizhou, Zhejiang China Guangdong Xinbao Electrical Appliances Holdings Co., Ltd. South Zhenghe Road, Leliu Town, Shunde District, Foshan, Guangdong China Yangzhou Juli Sports Articles Co., Ltd. Fudong Village, Xiaoji Town, Jiangdu District, Yangzhou, Jiangsu China Eyarn Lighting Ltd. Yaying Gang, Shixi Village, Shishan Town, Nanhai District, Foshan, Guangdong China Lipan Gift & Lighting Co., Ltd. No. 2 Guliao Road 3, Science Industrial Zone, Tangxia Town, Dongguan, Guangdong China Zhan Jiang Kang Nian Rubber Product Co., Ltd. No. 85 Middle Shen Chuan Road, Zhanjiang, Guangdong China Ansen Electronics Co. Ning Tau Administrative District, Qiao Tau Zhen, Dongguan, Guangdong China Changshu Tongrun Auto Accessory Co., Ltd. -

Plant List for VC54, North Lincolnshire
Plant List for Vice-county 54, North Lincolnshire 3 Vc61 SE TA 2 Vc63 1 SE TA SK NORTH LINCOLNSHIRE TF 9 8 Vc54 Vc56 7 6 5 Vc53 4 3 SK TF 6 7 8 9 1 2 3 4 5 6 Paul Kirby, 31/01/2017 Plant list for Vice-county 54, North Lincolnshire CONTENTS Introduction Page 1 - 50 Main Table 51 - 64 Summary Tables Red Listed taxa recorded between 2000 & 2017 51 Table 2 Threatened: Critically Endangered & Endangered 52 Table 3 Threatened: Vulnerable 53 Table 4 Near Threatened Nationally Rare & Scarce taxa recorded between 2000 & 2017 54 Table 5 Rare 55 - 56 Table 6 Scarce Vc54 Rare & Scarce taxa recorded between 2000 & 2017 57 - 59 Table 7 Rare 60 - 61 Table 8 Scarce Natives & Archaeophytes extinct & thought to be extinct in Vc54 62 - 64 Table 9 Extinct Plant list for Vice-county 54, North Lincolnshire The main table details all the Vascular Plant & Stonewort taxa with records on the MapMate botanical database for Vc54 at the end of January 2017. The table comprises: Column 1 Taxon and Authority 2 Common Name 3 Total number of records for the taxon on the database at 31/01/2017 4 Year of first record 5 Year of latest record 6 Number of hectads with records before 1/01/2000 7 Number of hectads with records between 1/01/2000 & 31/01/2017 8 Number of tetrads with records between 1/01/2000 & 31/01/2017 9 Comment & Conservation status of the taxon in Vc54 10 Conservation status of the taxon in the UK A hectad is a 10km. -

Supplementary Materials for the Article: a Running Start Or a Clean Slate
Supplementary Materials for the article: A running start or a clean slate? How a history of cooperation affects the ability of cities to cooperate on environmental governance Rui Mu1, * and Wouter Spekkink2 1 Dalian University of Technology, Faculty of Humanities and Social Sciences; [email protected] 2 The University of Manchester, Sustainable Consumption Institute; [email protected] * Correspondence: [email protected]; Tel.: +86-139-0411-9150 Appendix 1: Environmental Governance Actions in Beijing-Tianjin-Hebei Urban Agglomeration Event time Preorder Action Event Event name and description Actors involved (year-month-day) event no. type no. Part A: Joint actions at the agglomeration level MOEP NDRC Action plan for Beijing, Tianjin, Hebei and the surrounding areas to implement MOIIT 2013 9 17 FJA R1 air pollution control MOF MOHURD NEA BG TG HP MOEP NDRC Collaboration mechanism of air pollution control in Beijing, Tianjin, Hebei and 2013 10 23 R1 FJA MOIIT R2 the surrounding areas MOF MOHURD CMA NEA MOT BG Coordination office of air pollution control in Beijing, Tianjin, Hebei and the 2013 10 23 R2 FJA TG R3 surrounding areas HP 1 MOEP NDRC MOIIT MOF MOHURD CMA NEA MOT BG Clean Production Improvement Plan for Key Industrial Enterprises in Beijing, TG 2014 1 9 R1 FJA R4 Tianjin, Hebei and the Surrounding Areas HP MOIIT MOEP BTHAPCLG Regional air pollution joint prevention and control forum (the first meeting) 2014 3 3 R2, R3 IJA BEPA R5 was held by the Coordination Office. TEPA HEPA Coordination working unit was established for comprehensive atmospheric BTHAPCLG 2014 3 25 R3 FJA R6 pollution control. -

Simulation of Storm Surge Inundation Under Different Typhoon Intensity Scenarios: Case Study of Pingyang County, China
Nat. Hazards Earth Syst. Sci., 20, 2777–2790, 2020 https://doi.org/10.5194/nhess-20-2777-2020 © Author(s) 2020. This work is distributed under the Creative Commons Attribution 4.0 License. Simulation of storm surge inundation under different typhoon intensity scenarios: case study of Pingyang County, China Xianwu Shi1,2,4, Pubing Yu3, Zhixing Guo1, Zhilin Sun2, Fuyuan Chen2, Xiuguang Wu2, Wenlong Cheng2, and Jian Zeng2 1National Marine Hazard Mitigation Service, Beijing, 100194, China 2Ocean College, Zhejiang University, Hangzhou, 310058, China 3Zhejiang Institute of Hydraulics and Estuary, Hangzhou, 310020, China 4Key Laboratory of Coastal Disaster and Protection, Ministry of Education, Hohai University, Nanjing, 210098, China Correspondence: Jian Zeng ([email protected]) Received: 30 December 2019 – Discussion started: 5 February 2020 Revised: 7 September 2020 – Accepted: 13 September 2020 – Published: 21 October 2020 Abstract. China is one of the countries that is most seri- eas, which can be extremely destructive and can have a seri- ously affected by storm surges. In recent years, storm surges ous impact on surrounding areas (Sun et al., 2015). Storm in coastal areas of China have caused huge economic losses surges have occurred along much of China’s coast from and a large number of human casualties. Knowledge of the south to north (Gao et al., 2014). On average, approximately inundation range and water depth of storm surges under dif- nine typhoons make landfall over China annually (Shi et al., ferent typhoon intensities could assist predisaster risk assess- 2015), most of which cause storm surges. In 2018, storm ment and making evacuation plans, as well as provide deci- surge disasters caused coastal flooding in China that resulted sion support for responding to storm surges. -

SUPPLIER LIST OCTOBER 2019 Cotton on Group - Supplier List 2
SUPPLIER LIST OCTOBER 2019 Cotton On Group - Supplier List _2 REGION SUPPLIER NAME FACTORY NAME SUPPLIER ADDRESS PRODUCT TOTAL % OF % OF % OF TYPE WORKERS FEMALE MIGRANT TEMP WORKERS WORKERS WORKERS BANGLADESH APTECH DESIGN LTD DESIGNER FASHION LTD GOHAILBARI SHIMULIA APPAREL 3422 72% 0% 0% SAVAR DHAKA BANGLADESH APTECH DESIGN LTD Y-FRONTS ACCESSORIES (NEW HOUSE 24, WARD # 01, ARSHINAGOR MAIN ROAD APPAREL 24 0% 0% 0% LOCATION) ARSHINAGOR, KERANIGONJ DHAKA BANGLADESH BELAMY TEXTILES LTD BELAMY TEXTILES LTD KHOWAZ NAGAR, AZIMPARA APPAREL KARNAFULLY CHITTAGONG BANGLADESH BIG BOSS CORPORATION LTD (of aptech BIG BOSS CORPORATION LTD (NEW) APTECH INDUSTRIAL PARK APPAREL group) 30 SHARABO,KASIMPUR GAZIPUR BANGLADESH CLASSIC FASHIONS FOUR DESIGN PVT LTD (NEW) PLOT NO. B-201, 202, BSCIC HOSIERY I/E APPAREL 305 64% 0% 0% SHASONGAON, ENAYETNAGAR, FATULLAH NARAYANGANJ BANGLADESH CLASSIC FASHIONS FOUR DESIGN PVT LTD (TEMPORARY) PLOT NO. B327/328, BSCIC HOSIERY I/E APPAREL 380 60% 0% 0% SHASONGAON, ENAYETNAGAR, FATULLAH NARAYANGANJ BANGLADESH IMPRESS NEWTEX COMPOSITE TEXTILES B2B EXCELLANCE LTD MIRZAPUR PURBAPARA, APPAREL 1355 76% 0% 0% LTD 8 NO. MIRZAPUR MOUZA, GAZIPUR SADAR,GAZIPUR BANGLADESH IMPRESS NEWTEX COMPOSITE TEXTILES IMPRESS NEWTEX COMPOSITE TEXTILES GORAI INDUSTRIAL AREA APPAREL 2124 56% 0% 0% LTD LTD MIRZAPUR TANGAIL BANGLADESH IRIS FABRICS LIMITED IRIS FABRICS LIMITED ZIRANIBAZAR APPAREL 2984 47% 0% 0% KASHIMPUT, JOYDEVPUR GAZIPUR BANGLADESH JERICHO IMEX JERICHO IMEX LTD MONTREE BARI ROAD APPAREL 700 62% 0% 0% SOUTH SHANLA, SHANLA BAZAR -
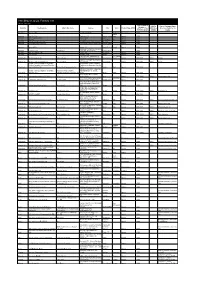
Levi Strauss & Co. Factory List
Levi Strauss & Co. Factory List Published : August 2020 Total Number of LS&Co. Parent Company Name Employees Country Factory name Alternative Name Address City State Product Type (TOE) Initiatives (Licensee factories are (Workers, Staff, (WWB) blank) Contract Staff) Argentina Accecuer SA Juan Zanella 4656 Caseros Dong Nai Accessories <1000 Tanmil Argentina Best Sox S.A. Charlone 1446 Capital Federal Nadu Apparel <1000 Argentina Estex Argentina S.R.L. Superi, 3530 Caba Apparel <1000 Argentina Gitti SRL Italia 4043 Mar del Plata TN Apparel <1000 Argentina Manufactura Arrecifes S.A. Ruta Nacional 8, Kilometro 178 Arrecifes Apparel <1000 Argentina Procesadora Serviconf SRL Gobernardor Ramon Castro 4765 Vicente Lopez Apparel <1000 Argentina Spring S.R.L. Darwin, 173 Capital Federal Punjab Apparel <1000 Asamblea (101) #536, Villa Lynch Argentina TEXINTER S.A. Texinter S.A. B1672AIB, Buenos Aires Buenos Aires <1000 Argentina Underwear M&S, S.R.L Levalle 449 Avellaneda Apparel <1000 Argentina Vira Offis S.A. Virasoro, 3570 Rosario Apparel <1000 Plot # 246-249, Adamjee EPZ, Rio Grande Bangladesh Ananta Apparels Ltd. Nazmul Hoque Shiddirgonj, Narayangonj-1431 Narayangonj do Sul Apparel 1000-5000 WWB Ananta Nayabari, Kanchpur, Sonargaon, Bangladesh Ananta Denim Technology Ltd. Md. Ziaul Alam Narayangonj. Narayanganj Apparel 1000-5000 WWB Ananta Ayesha Clothing Company Ltd (Ayesha Bangobandhu Road, Tongabari, Clothing Company Ltd,Hamza Trims Ltd, Gazirchat Alia Madrasha, Ashulia, Bangladesh Hamza Clothing Ltd) Ayesha Clothing Company Ltd( Dhaka Dhaka Sindh Apparel 1000-5000 Jamgora, Post Office : Gazirchat Ayesha Clothing Company Ltd (Ayesha Ayesha Clothing Company Alia Madrasha, P.S : Savar, Bangladesh Washing Ltd.) Ltd(Unit-1)d (Ayesha Washing Ltd) Dhaka Dhaka Sindh Apparel 1000-5000 Khejur Bagan, Bara Ashulia, Bangladesh Cosmopolitan Industries PVT Ltd CIPL Savar, Dhaka Savar Apparel 5000-10000 WWB Epic Designers Ltd 1612, South Salna, Salna Bazar, TAMILNAD Bangladesh Cutting Edge Industries Ltd. -

SUPPLIER LIST AUGUST 2019 Cotton on Group - Supplier List 2
SUPPLIER LIST AUGUST 2019 Cotton On Group - Supplier List _2 COUNTRY FACTORY NAME SUPPLIER ADDRESS STAGE TOTAL % OF % OF % OF TEMP WORKERS FEMALE MIGRANT WORKER WORKERS WORKER CHINA NINGBO FORTUNE INTERNATIONAL TRADE CO LTD RM 805-8078 728 LANE SONGJIANG EAST ROAD SUP YINZHOU NINGBO, ZHEJIANG CHINA NINGBO QIANZHEN CLOTHES CO LTD OUCHI VILLAGE CMT 104 64% 75% 6% GULIN TOWN, HAISHU DISTRICT NINGBO, ZHEJIANG CHINA XIANGSHAN YUFA KNITTING LTD NO.35 ZHENYIN RD, JUEXI STREET CMT 57 60% 88% 12% XIANGSHAN COUNTY NINGBO CITY, ZHEJIANG CHINA SUNROSE INTERNATIONAL CO LTD ROOM 22/2 227 JINMEI BUILDING NO 58 LANE 136 SUP SHUNDE ROAD, HAISHU DISTRICT NINGBO, ZHEJIANG CHINA NINGBO HAISHU WANQIANYAO TEXTILE CO LTD NO 197 SAN SAN ROAD CMT 26 62% 85% 0% WANGCHAN INDUSTRIAL ZONE NINGBO, ZHEJIANG CHINA ZHUJI JUNHANG SOCKS FACTORY DAMO VILLAGE LUXI NEW VILLAGE CMT 73 38% 66% 0% ZHUJI CITY ZHEJIANG CHINA SKYLEAD INDUSTRY CO LIMITED LAIMEI INDUSTRIAL PARK SUP CHENGHAI DISTRICT, SHANTOU CITY GUANGDONG CHINA CHUANGXIANG TOYS LIMITED LAIMEI INDUSTRIAL PARK CMT 49 33% 90% 0% CHENGHAI DISTRICT SHANTOU, GUANGDONG CHINA NINGBO ODESUN STATIONERY & GIFT CO LTD TONGJIA VILLAGE, PANHUO INDUSTRIAL ZONE SUP YINZHOU DISTRICT NINGBO CITY, ZHEJIANG CHINA NINGBO ODESUN STATIONERY & GIFT CO LTD TONGJIA VILLAGE, PANHUO INDUSTRIAL ZONE CMT YINZHOU DISTRICT NINGBO CITY, ZHEJIANG CHINA NINGBO WORTH INTERNATIONAL TRADE CO LTD RM. 1902 BUILDING B, CROWN WORLD TRADE PLAZA SUP NO. 1 LANE 28 BAIZHANG EAST ROAD NINGBO ZHEJIANG CHINA NINGHAI YUEMING METAL PRODUCT CO LTD NO. 5 HONGTA ROAD -

The Order of Local Things: Popular Politics and Religion in Modern
The Order of Local Things: Popular Politics and Religion in Modern Wenzhou, 1840-1940 By Shih-Chieh Lo B.A., National Chung Cheng University, 1997 M.A., National Tsing Hua University, 2000 A.M., Brown University, 2005 Submitted in Partial Fulfillment for the Degree of Doctor of Philosophy in the Department of History at Brown University PROVIDENCE, RHODE ISLAND May 2010 © Copyright 2010 by Shih-Chieh Lo ii This dissertation by Shih-Chieh Lo is accepted in its present form by the Department of History as satisfying the dissertation requirement for the degree of Doctor of Philosophy. Date_____________ ________________________ Mark Swislocki, Advisor Recommendation to the Graduate Council Date_____________ __________________________ Michael Szonyi, Reader Date_____________ __________________________ Mark Swislocki, Reader Date_____________ __________________________ Richard Davis, Reader Approved by the Graduate Council Date______________ ___________________________ Sheila Bonde, Dean of the Graduate School iii Roger, Shih-Chieh Lo (C. J. Low) Date of Birth : August 15, 1974 Place of Birth : Taichung County, Taiwan Education Brown University- Providence, Rhode Island Ph. D in History (May 2010) Brown University - Providence, Rhode Island A. M., History (May 2005) National Tsing Hua University- Hsinchu, Taiwan Master of Arts (June 2000) National Chung-Cheng University - Chaiyi, Taiwan Bachelor of Arts (June 1997) Publications: “地方神明如何平定叛亂:楊府君與溫州地方政治 (1830-1860).” (How a local deity pacified Rebellion: Yangfu Jun and Wenzhou local politics, 1830-1860) Journal of Wenzhou University. Social Sciences 溫州大學學報 社會科學版, Vol. 23, No.2 (March, 2010): 1-13. “ 略論清同治年間台灣戴潮春案與天地會之關係 Was the Dai Chaochun Incident a Triad Rebellion?” Journal of Chinese Ritual, Theatre and Folklore 民俗曲藝 Vol. 138 (December, 2002): 279-303. “ 試探清代台灣的地方精英與地方社會: 以同治年間的戴潮春案為討論中心 Preliminary Understandings of Local Elites and Local Society in Qing Taiwan: A Case Study of the Dai Chaochun Rebellion”. -

Large-Scale Natural Disaster Risk Scenario Analysis: a Case Study of Wenzhou City, China
Nat Hazards (2012) 60:1287–1298 DOI 10.1007/s11069-011-9909-2 ORIGINAL PAPER Large-scale natural disaster risk scenario analysis: a case study of Wenzhou City, China Yaolong Liu • Zhenlou Chen • Jun Wang • Beibei Hu • Mingwu Ye • Shiyuan Xu Received: 17 September 2010 / Accepted: 16 July 2011 / Published online: 14 August 2011 Ó Springer Science+Business Media B.V. 2011 Abstract Based on the analysis and calculation of the hazard intensity of typhoon rainstorms and floods as well as the vulnerability of flood receptors and the possibility of great losses, risk scenarios are proposed and presented in Wenzhou City, Zhejiang Prov- ince, China, using the Pearson-III model and ArcGIS spatial analyst tools. Results indicate that the elements of risk scenarios include time–space scenarios, disaster scenarios, and man-made scenarios. Ten-year and 100-year typhoon rainstorms and flood hazard areas are mainly concentrated in the coastal areas of Wenzhou City. The average rainfall across a 100-year frequency is 450 mm. The extreme water depth of a 100-year flood is 600 mm. High-vulnerability areas are located in Yueqing, Pingyang, Cangnan, and Wencheng counties. The average loss rate of a 100-year flood is more than 50%. The greatest possible loss of floods shows an obvious concentration-diffusion situation. There is an area of about 20–25% flood loss of 6–24 million Yuan RMB/km2 in the Lucheng, Longwan and Ouhai districts. The average loss of a 100-year flood is 12 million Yuan RMB/km2, and extreme loss reaches 49.33 million Yuan RMB/km2.