Hereditary Multiple Osteochondromas
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Familial Nephropathy and Multiple Exostoses with Exostosin-1 (EXT1) Gene Mutation
PATHOPHYSIOLOGY of the RENAL BIOPSY www.jasn.org Familial Nephropathy and Multiple Exostoses With Exostosin-1 (EXT1) Gene Mutation Ian S. D. Roberts* and Jonathan M. Gleadle† *Department of Cellular Pathology, John Radcliffe Hospital, Headley Way, Headington, Oxford, United Kingdom; and †Renal Unit, Level 6, Flinders Medical Centre, Bedford Park, South Australia, Australia ABSTRACT Glomerular deposition of fibrillar collagen is a characteristic finding of genetically mained in remission with trace protein- distinct conditions, including nail-patella syndrome and collagen type III glomeru- uria until cyclosporine was stopped 3.5 lopathy. A case of familial nephropathy in which steroid-sensitive nephrotic syn- yr later. Six months after this, she suf- drome and glomerular deposits of fibrillar collagen are associated with multiple fered another relapse of nephrotic exostoses due to mutation of the EXT1 gene is described. This gene encodes a syndrome that responded to 60 mg pred- glycosyltransferase required for synthesis of heparan sulfate glycosaminoglycans. nisolone and reintroduction of cyclo- There is deficiency of heparan sulfate and perlecan, together with accumulation of sporine. After a further relapse 18 mo collagens, in the matrix of EXT1-associated osteochondromas. Similar glomerular later and because of the development of basement membrane abnormalities could offer an explanation for both the renal adverse corticosteroid effects, she was ultrastructural changes and steroid-sensitive nephrotic syndrome. treated with a 2-mo course of cyclophos- phamide (2.5 mg/kg, orally). Ten years J Am Soc Nephrol 19: 450–453, 2008. doi: 10.1681/ASN.2007080842 after her initial presentation, she remains in full remission and off steroids. Renal function has remained normal through- A 37-yr-old woman presented with the history of renal disease and hearing im- out with a current serum creatinine of nephrotic syndrome. -

CRISPR Screening of Porcine Sgrna Library Identifies Host Factors
ARTICLE https://doi.org/10.1038/s41467-020-18936-1 OPEN CRISPR screening of porcine sgRNA library identifies host factors associated with Japanese encephalitis virus replication Changzhi Zhao1,5, Hailong Liu1,5, Tianhe Xiao1,5, Zichang Wang1, Xiongwei Nie1, Xinyun Li1,2, Ping Qian2,3, Liuxing Qin3, Xiaosong Han1, Jinfu Zhang1, Jinxue Ruan1, Mengjin Zhu1,2, Yi-Liang Miao 1,2, Bo Zuo1,2, ✉ ✉ Kui Yang4, Shengsong Xie 1,2 & Shuhong Zhao 1,2 1234567890():,; Japanese encephalitis virus (JEV) is a mosquito-borne zoonotic flavivirus that causes ence- phalitis and reproductive disorders in mammalian species. However, the host factors critical for its entry, replication, and assembly are poorly understood. Here, we design a porcine genome-scale CRISPR/Cas9 knockout (PigGeCKO) library containing 85,674 single guide RNAs targeting 17,743 protein-coding genes, 11,053 long ncRNAs, and 551 microRNAs. Subsequently, we use the PigGeCKO library to identify key host factors facilitating JEV infection in porcine cells. Several previously unreported genes required for JEV infection are highly enriched post-JEV selection. We conduct follow-up studies to verify the dependency of JEV on these genes, and identify functional contributions for six of the many candidate JEV- related host genes, including EMC3 and CALR. Additionally, we identify that four genes associated with heparan sulfate proteoglycans (HSPGs) metabolism, specifically those responsible for HSPGs sulfurylation, facilitate JEV entry into porcine cells. Thus, beyond our development of the largest CRISPR-based functional genomic screening platform for pig research to date, this study identifies multiple potentially vulnerable targets for the devel- opment of medical and breeding technologies to treat and prevent diseases caused by JEV. -

Selectin Ligand Sialyl-Lewis X Antigen Drives Metastasis of Hormone-Dependent Breast Cancers
Published OnlineFirst October 24, 2011; DOI: 10.1158/0008-5472.CAN-11-1139 Cancer Tumor and Stem Cell Biology Research Selectin Ligand Sialyl-Lewis x Antigen Drives Metastasis of Hormone-Dependent Breast Cancers Sylvain Julien1, Aleksandar Ivetic2, Anita Grigoriadis3, Ding QiZe1, Brian Burford1, Daisy Sproviero1, Gianfranco Picco1, Cheryl Gillett4, Suzanne L. Papp5, Lana Schaffer5, Andrew Tutt3, Joyce Taylor-Papadimitriou1, Sarah E. Pinder4, and Joy M. Burchell1 Abstract The glycome acts as an essential interface between cells and the surrounding microenvironment. However, changes in glycosylation occur in nearly all breast cancers, which can alter this interaction. Here, we report that profiles of glycosylation vary between ER-positive and ER-negative breast cancers. We found that genes involved in the synthesis of sialyl-Lewis x (sLex; FUT3, FUT4, and ST3GAL6) are significantly increased in estrogen receptor alpha-negative (ER-negative) tumors compared with ER-positive ones. SLex expression had no influence on the survival of patients whether they had ER-negative or ER-positive tumors. However, high expression of sLex in ER- positive tumors was correlated with metastasis to the bone where sLex receptor E-selectin is constitutively expressed. The ER-positive ZR-75-1 and the ER-negative BT20 cell lines both express sLex but only ZR-75-1 cells could adhere to activated endothelial cells under dynamic flow conditions in a sLex and E-selectin–dependent manner. Moreover, L/P-selectins bound strongly to ER-negative MDA-MB-231 and BT-20 cell lines in a heparan sulfate (HS)–dependent manner that was independent of sLex expression. Expression of glycosylation genes involved in heparan biosynthesis (EXT1 and HS3ST1) was increased in ER-negative tumors. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

ZNF263 Is a Transcriptional Regulator of Heparin and Heparan Sulfate Biosynthesis
ZNF263 is a transcriptional regulator of heparin and heparan sulfate biosynthesis Ryan J. Weissa,1, Philipp N. Spahnb,1, Alejandro Gómez Toledoa, Austin W. T. Chiangb, Benjamin P. Kellmanb,JingLia, Christopher Bennerc, Christopher K. Glassa,c,PhilipL.S.M.Gordtsc,d,NathanE.Lewisb,d,e,2, and Jeffrey D. Eskoa,d,2,3 aDepartment of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093-0687; bDepartment of Pediatrics, University of California San Diego, La Jolla, CA 92093-0760; cDepartment of Medicine, University of California San Diego, La Jolla, CA 92093-0687; dGlycobiology Research and Training Center, University of California San Diego, La Jolla, CA 92093-0687; and eDepartment of Bioengineering, University of California San Diego, La Jolla, CA 92093-0687 Edited by Tadatsugu Taniguchi, University of Tokyo, Meguro-ku, Japan, and approved March 9, 2020 (received for review December 2, 2019) Heparin is the most widely prescribed biopharmaceutical in pro- inactivate thrombin and Factor Xa, which accounts for its potent duction globally. Its potent anticoagulant activity and safety makes anticoagulant activity (4). it the drug of choice for preventing deep vein thrombosis and In 2008, the US Food and Drug Administration issued a major pulmonary embolism. In 2008, adulterated material was intro- recall of pharmaceutical heparin due to contamination of the duced into the heparin supply chain, resulting in several hundred raw heparin stock imported from China. This crisis prompted deaths and demonstrating the need for alternate sources of heparin. new guidelines for monitoring the purity of heparin, but the Heparin is a fractionated form of heparan sulfate derived from feedstock remains vulnerable to natural variation, susceptibility animal sources, predominantly from connective tissue mast cells in of the pig population to infectious agents, and potential con- pig mucosa. -

Viruses Like Sugars: How to Assess Glycan Involvement in Viral Attachment
microorganisms Review Viruses Like Sugars: How to Assess Glycan Involvement in Viral Attachment Gregory Mathez and Valeria Cagno * Institute of Microbiology, Lausanne University Hospital, University of Lausanne, 1011 Lausanne, Switzerland; [email protected] * Correspondence: [email protected] Abstract: The first step of viral infection requires interaction with the host cell. Before finding the specific receptor that triggers entry, the majority of viruses interact with the glycocalyx. Identifying the carbohydrates that are specifically recognized by different viruses is important both for assessing the cellular tropism and for identifying new antiviral targets. Advances in the tools available for studying glycan–protein interactions have made it possible to identify them more rapidly; however, it is important to recognize the limitations of these methods in order to draw relevant conclusions. Here, we review different techniques: genetic screening, glycan arrays, enzymatic and pharmacological approaches, and surface plasmon resonance. We then detail the glycan interactions of enterovirus D68 and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), highlighting the aspects that need further clarification. Keywords: attachment receptor; viruses; glycan; sialic acid; heparan sulfate; HBGA; SARS-CoV-2; EV-D68 Citation: Mathez, G.; Cagno, V. Viruses Like Sugars: How to Assess 1. Introduction Glycan Involvement in Viral This review focuses on methods for assessing the involvement of carbohydrates in Attachment. Microorganisms 2021, 9, viral attachment and entry into the host cell. Viruses often bind to entry receptors that are 1238. https://doi.org/10.3390/ not abundant on the cell surface; to increase their chances of finding them, they initially microorganisms9061238 bind to attachment receptors comprising carbohydrates that are more widely expressed. -
Drosophila and Human Transcriptomic Data Mining Provides Evidence for Therapeutic
Drosophila and human transcriptomic data mining provides evidence for therapeutic mechanism of pentylenetetrazole in Down syndrome Author Abhay Sharma Institute of Genomics and Integrative Biology Council of Scientific and Industrial Research Delhi University Campus, Mall Road Delhi 110007, India Tel: +91-11-27666156, Fax: +91-11-27662407 Email: [email protected] Nature Precedings : hdl:10101/npre.2010.4330.1 Posted 5 Apr 2010 Running head: Pentylenetetrazole mechanism in Down syndrome 1 Abstract Pentylenetetrazole (PTZ) has recently been found to ameliorate cognitive impairment in rodent models of Down syndrome (DS). The mechanism underlying PTZ’s therapeutic effect is however not clear. Microarray profiling has previously reported differential expression of genes in DS. No mammalian transcriptomic data on PTZ treatment however exists. Nevertheless, a Drosophila model inspired by rodent models of PTZ induced kindling plasticity has recently been described. Microarray profiling has shown PTZ’s downregulatory effect on gene expression in fly heads. In a comparative transcriptomics approach, I have analyzed the available microarray data in order to identify potential mechanism of PTZ action in DS. I find that transcriptomic correlates of chronic PTZ in Drosophila and DS counteract each other. A significant enrichment is observed between PTZ downregulated and DS upregulated genes, and a significant depletion between PTZ downregulated and DS dowwnregulated genes. Further, the common genes in PTZ Nature Precedings : hdl:10101/npre.2010.4330.1 Posted 5 Apr 2010 downregulated and DS upregulated sets show enrichment for MAP kinase pathway. My analysis suggests that downregulation of MAP kinase pathway may mediate therapeutic effect of PTZ in DS. Existing evidence implicating MAP kinase pathway in DS supports this observation. -

Drug Metabolism Determines Resistance of Colorectal Cancer to Resorcinol-Based HSP90 Inhibitors
Drug Metabolism Determines Resistance of Colorectal Cancer to Resorcinol-Based HSP90 Inhibitors Dissertation for the award of the degree “Doctor rerum naturalium” (Dr. rer. nat.) in the “Biology of Cells” Program at the Georg August University Göttingen, Faculty of Biology submitted by Hannes Landmann born in Hannover, Germany Göttingen, August 2014 Thesis Committee Prof. Dr. Matthias Dobbelstein Institute for Molecular Oncology, Faculty of Medicine Prof. Dr. Heidi Hahn Tumor Genetics, Department of Human Genetics, Faculty of Medicine Prof. Dr. Dieter Kube Department for Hematology and Oncology, Faculty of Medicine Members of the Examination Board Referee: Prof. Dr. Matthias Dobbelstein, Institute for Molecular Oncology 2nd Referee: Prof. Dr. Heidi Hahn, Tumor Genetics, Department of Human Genetics Further members of the Examination Board Prof. Dr. Dieter Kube Department for Hematology and Oncology, Faculty of Medicine PD Dr. Halyna Shcherbata Research Group Gene Expression and Signaling, Max Planck Institute for Biophysical Chemistry Prof. Dr. Ralph Kehlenbach Department of Molecular Biology, Faculty of Biochemistry PD Dr. Wilfried Kramer Department for Molecular Genetics, Institute for Microbiology and Genetics Date of oral examination: 19th of September 2014 AFFIDAVIT Herewith I declare that I prepared the PhD thesis “Drug Metabolism Determines Resistance of Colorectal Cancer to Resorcinol-Based HSP90 Inhibitors” on my own with no other aids and sources than quoted. _____________________ Hannes Landmann, Göttingen in August 2014 The findings in this work were accepted for publication in the open access journal Cell Death and Disease: Landmann, H., Proia, D.A., He, S., Ogawa, L.S., Kramer, F., Beißbarth, T., Grade, M., Gaedcke, J., Ghadimi, M., Moll, U. -

A Mouse Model of Chondrocyte-Specific Somatic
A mouse model of chondrocyte-specific somatic mutation reveals a role for Ext1 loss of heterozygosity in multiple hereditary exostoses Kazu Matsumotoa, Fumitoshi Iriea, Susan Mackemb, and Yu Yamaguchia,1 aSanford Children’s Health Research Center, Sanford-Burnham Medical Research Institute, La Jolla, CA 92037; and bCancer and Developmental Biology Laboratory, National Cancer Institute, Frederick, MD 21702 Edited* by Erkki Ruoslahti, University of California, Santa Barbara, CA, and approved May 12, 2010 (received for review December 23, 2009) Multiple hereditary exostoses (MHE) is one of the most common The current consensus is that the copolymerase activity resides skeletal dysplasias, exhibiting the formation of multiple cartilage- in the EXT1 protein, and that the association of the EXT2 capped bony protrusions (osteochondroma) and characteristic protein with the EXT1 protein is necessary for the proper lo- bone deformities. Individuals with MHE carry heterozygous loss- calization of EXT1 in the Golgi apparatus (14, 15). Genetic of-function mutations in Ext1 or Ext2, genes which together encode ablation of either gene results in essentially complete abrogation an enzyme essential for heparan sulfate synthesis. Despite the of HS production in cells and tissues (16, 17). The majority of identification of causative genes, the pathogenesis of MHE remains cases of MHE carry frameshift or missense mutations in EXT1 or unclear, especially with regard to whether osteochondroma results EXT2 (18, 19). from loss of heterozygosity of the Ext genes. Hampering elucida- Despite the unambiguous identification of causative genes and +/− +/− tion of the pathogenic mechanism of MHE, both Ext1 and Ext2 their function, the pathogenic mechanism of MHE remains heterozygous mutant mice, which mimic the genetic status of hu- elusive. -

MDA-MB-231 Breast Cancer Cell Viability, Motility and Matrix
Glycoconj J DOI 10.1007/s10719-016-9735-6 ORIGINAL ARTICLE MDA-MB-231 breast cancer cell viability, motility and matrix adhesion are regulated by a complex interplay of heparan sulfate, chondroitin−/dermatan sulfate and hyaluronan biosynthesis Manuela Viola1 & Kathrin Brüggemann2 & Evgenia Karousou1 & Ilaria Caon1 & Elena Caravà1 & Davide Vigetti1 & Burkhard Greve3 & Christian Stock4,5 & Giancarlo De Luca1 & Alberto Passi1 & Martin Götte2 Received: 30 June 2016 /Revised: 23 September 2016 /Accepted: 28 September 2016 # Springer Science+Business Media New York 2016 Abstract Proteoglycans and glycosaminoglycans modulate parameters were unchanged in EXT1-silenced cells. numerous cellular processes relevant to tumour progression, Importantly, these changes were associated with a decreased including cell proliferation, cell-matrix interactions, cell mo- expression of syndecan-1, decreased signalling response to tility and invasive growth. Among the glycosaminoglycans HGF and an increase in the synthesis of hyaluronan, due to with a well-documented role in tumour progression are hepa- an upregulation of the hyaluronan synthases HAS2 and ran sulphate, chondroitin/dermatan sulphate and hyaluronic HAS3. Interestingly, EXT1-depleted cells showed a downreg- acid/hyaluronan. While the mode of biosynthesis differs for ulation of the UDP-sugar transporter SLC35D1, whereas sulphated glycosaminoglycans, which are synthesised in the SLC35D2 was downregulated in β4GalT7-depleted cells, in- ER and Golgi compartments, and hyaluronan, which is syn- dicating an intricate regulatory network that connects all gly- thesized at the plasma membrane, these polysaccharides par- cosaminoglycans synthesis. The results of our in vitro study tially compete for common substrates. In this study, we suggest that a modulation of breast cancer cell behaviour via employed a siRNA knockdown approach for heparan interference with heparan sulphate biosynthesis may result in sulphate (EXT1) and heparan/chondroitin/dermatan a compensatory upregulation of hyaluronan biosynthesis. -
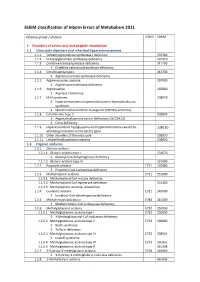
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2. -

Multiplexed Surrogate Analysis of Glycotransferase Activity in Whole Biospecimens † † ‡ Chad R
Article pubs.acs.org/ac Multiplexed Surrogate Analysis of Glycotransferase Activity in Whole Biospecimens † † ‡ Chad R. Borges, ,* Douglas S. Rehder, and Paolo Boffetta † Molecular Biomarkers Unit, The Biodesign Institute at Arizona State University, Tempe, Arizona 85287, United States ‡ Institute for Translational Epidemiology and Tisch Cancer Institute, Mount Sinai School of Medicine, New York, New York 10029, United States *S Supporting Information ABSTRACT: Dysregulated glycotransferase enzymes in can- cer cells produce aberrant glycanssome of which can help facilitate metastases. Within a cell, individual glycotransferases promiscuously help to construct dozens of unique glycan structures, making it difficult to comprehensively track their activity in biospecimensespecially where they are absent or inactive. Here, we describe an approach to deconstruct glycans in whole biospecimens then analytically pool together resulting monosaccharide-and-linkage-specific degradation products (“glycan nodes”) that directly represent the activities of specific glycotransferases. To implement this concept, a reproducible, relative quantitation-based glycan methylation analysis methodology was developed that simultaneously captures information from N-, O-, and lipid linked glycans and is compatible with whole biofluids and homogenized tissues; in total, over 30 different glycan nodes are detectable per gas chromatography−mass spectrometry (GC-MS) run. Numerous nonliver organ cancers are known to induce the production of abnormally glycosylated serum proteins. Thus, following analytical validation, in blood plasma, the technique was applied to a group of 59 lung cancer patient plasma samples and age/gender/ smoking-status-matched non-neoplastic controls from the Lung Cancer in Central and Eastern Europe (CEE) study to gauge the clinical utility of the approach toward the detection of lung cancer.