Mouse Ap2a2 Conditional Knockout Project (CRISPR/Cas9)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

AP2A2 Antibody Cat
AP2A2 Antibody Cat. No.: 63-513 AP2A2 Antibody Formalin-fixed and paraffin-embedded human Flow cytometric analysis of HepG2 cells using AP2A2 hepatocarcinoma with AP2A2 Antibody , which was Antibody (bottom histogram) compared to a negative peroxidase-conjugated to the secondary antibody, control cell (top histogram). FITC-conjugated goat-anti- followed by DAB staining. rabbit secondary antibodies were used for the analysis. Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human This AP2A2 antibody is generated from rabbits immunized with a KLH conjugated IMMUNOGEN: synthetic peptide between 610-637 amino acids from the Central region of human AP2A2. TESTED APPLICATIONS: Flow, IHC-P, WB For WB starting dilution is: 1:1000 APPLICATIONS: For IHC-P starting dilution is: 1:10~50 For FACS starting dilution is: 1:10~50 September 25, 2021 1 https://www.prosci-inc.com/ap2a2-antibody-63-513.html PREDICTED MOLECULAR 104 kDa WEIGHT: Properties This antibody is purified through a protein A column, followed by peptide affinity PURIFICATION: purification. CLONALITY: Polyclonal ISOTYPE: Rabbit Ig CONJUGATE: Unconjugated PHYSICAL STATE: Liquid BUFFER: Supplied in PBS with 0.09% (W/V) sodium azide. CONCENTRATION: batch dependent Store at 4˚C for three months and -20˚C, stable for up to one year. As with all antibodies STORAGE CONDITIONS: care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. Additional Info OFFICIAL SYMBOL: AP2A2 AP-2 complex subunit alpha-2, 100 kDa coated vesicle -

Supplemental Information
Supplemental information Dissection of the genomic structure of the miR-183/96/182 gene. Previously, we showed that the miR-183/96/182 cluster is an intergenic miRNA cluster, located in a ~60-kb interval between the genes encoding nuclear respiratory factor-1 (Nrf1) and ubiquitin-conjugating enzyme E2H (Ube2h) on mouse chr6qA3.3 (1). To start to uncover the genomic structure of the miR- 183/96/182 gene, we first studied genomic features around miR-183/96/182 in the UCSC genome browser (http://genome.UCSC.edu/), and identified two CpG islands 3.4-6.5 kb 5’ of pre-miR-183, the most 5’ miRNA of the cluster (Fig. 1A; Fig. S1 and Seq. S1). A cDNA clone, AK044220, located at 3.2-4.6 kb 5’ to pre-miR-183, encompasses the second CpG island (Fig. 1A; Fig. S1). We hypothesized that this cDNA clone was derived from 5’ exon(s) of the primary transcript of the miR-183/96/182 gene, as CpG islands are often associated with promoters (2). Supporting this hypothesis, multiple expressed sequences detected by gene-trap clones, including clone D016D06 (3, 4), were co-localized with the cDNA clone AK044220 (Fig. 1A; Fig. S1). Clone D016D06, deposited by the German GeneTrap Consortium (GGTC) (http://tikus.gsf.de) (3, 4), was derived from insertion of a retroviral construct, rFlpROSAβgeo in 129S2 ES cells (Fig. 1A and C). The rFlpROSAβgeo construct carries a promoterless reporter gene, the β−geo cassette - an in-frame fusion of the β-galactosidase and neomycin resistance (Neor) gene (5), with a splicing acceptor (SA) immediately upstream, and a polyA signal downstream of the β−geo cassette (Fig. -

Ykt6 Membrane-To-Cytosol Cycling Regulates Exosomal Wnt Secretion
bioRxiv preprint doi: https://doi.org/10.1101/485565; this version posted December 3, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Ykt6 membrane-to-cytosol cycling regulates exosomal Wnt secretion Karen Linnemannstöns1,2, Pradhipa Karuna1,2, Leonie Witte1,2, Jeanette Kittel1,2, Adi Danieli1,2, Denise Müller1,2, Lena Nitsch1,2, Mona Honemann-Capito1,2, Ferdinand Grawe3,4, Andreas Wodarz3,4 and Julia Christina Gross1,2* Affiliations: 1Hematology and Oncology, University Medical Center Goettingen, Goettingen, Germany. 2Developmental Biochemistry, University Medical Center Goettingen, Goettingen, Germany. 3Molecular Cell Biology, Institute I for Anatomy, University of Cologne Medical School, Cologne, Germany 4Cluster of Excellence-Cellular Stress Response in Aging-Associated Diseases (CECAD), Cologne, Germany *Correspondence: Dr. Julia Christina Gross, Hematology and Oncology/Developmental Biochemistry, University Medical Center Goettingen, Justus-von-Liebig Weg 11, 37077 Goettingen Germany Abstract Protein trafficking in the secretory pathway, for example the secretion of Wnt proteins, requires tight regulation. These ligands activate Wnt signaling pathways and are crucially involved in development and disease. Wnt is transported to the plasma membrane by its cargo receptor Evi, where Wnt/Evi complexes are endocytosed and sorted onto exosomes for long-range secretion. However, the trafficking steps within the endosomal compartment are not fully understood. The promiscuous SNARE Ykt6 folds into an auto-inhibiting conformation in the cytosol, but a portion associates with membranes by its farnesylated and palmitoylated C-terminus. Here, we demonstrate that membrane detachment of Ykt6 is essential for exosomal Wnt secretion. -

Integrating Protein Copy Numbers with Interaction Networks to Quantify Stoichiometry in Mammalian Endocytosis
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.29.361196; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Integrating protein copy numbers with interaction networks to quantify stoichiometry in mammalian endocytosis Daisy Duan1, Meretta Hanson1, David O. Holland2, Margaret E Johnson1* 1TC Jenkins Department of Biophysics, Johns Hopkins University, 3400 N Charles St, Baltimore, MD 21218. 2NIH, Bethesda, MD, 20892. *Corresponding Author: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.10.29.361196; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Abstract Proteins that drive processes like clathrin-mediated endocytosis (CME) are expressed at various copy numbers within a cell, from hundreds (e.g. auxilin) to millions (e.g. clathrin). Between cell types with identical genomes, copy numbers further vary significantly both in absolute and relative abundance. These variations contain essential information about each protein’s function, but how significant are these variations and how can they be quantified to infer useful functional behavior? Here, we address this by quantifying the stoichiometry of proteins involved in the CME network. We find robust trends across three cell types in proteins that are sub- vs super-stoichiometric in terms of protein function, network topology (e.g. -

Supplementary Table 8. Cpcp PPI Network Details for Significantly Changed Proteins, As Identified in 3.2, Underlying Each of the Five Functional Domains
Supplementary Table 8. cPCP PPI network details for significantly changed proteins, as identified in 3.2, underlying each of the five functional domains. The network nodes represent each significant protein, followed by the list of interactors. Note that identifiers were converted to gene names to facilitate PPI database queries. Functional Domain Node Interactors Development and Park7 Rack1 differentiation Kcnma1 Atp6v1a Ywhae Ywhaz Pgls Hsd3b7 Development and Prdx6 Ncoa3 differentiation Pla2g4a Sufu Ncf2 Gstp1 Grin2b Ywhae Pgls Hsd3b7 Development and Atp1a2 Kcnma1 differentiation Vamp2 Development and Cntn1 Prnp differentiation Ywhaz Clstn1 Dlg4 App Ywhae Ywhab Development and Rac1 Pak1 differentiation Cdc42 Rhoa Dlg4 Ctnnb1 Mapk9 Mapk8 Pik3cb Sod1 Rrad Epb41l2 Nono Ltbp1 Evi5 Rbm39 Aplp2 Smurf2 Grin1 Grin2b Xiap Chn2 Cav1 Cybb Pgls Ywhae Development and Hbb-b1 Atp5b differentiation Hba Kcnma1 Got1 Aldoa Ywhaz Pgls Hsd3b4 Hsd3b7 Ywhae Development and Myh6 Mybpc3 differentiation Prkce Ywhae Development and Amph Capn2 differentiation Ap2a2 Dnm1 Dnm3 Dnm2 Atp6v1a Ywhab Development and Dnm3 Bin1 differentiation Amph Pacsin1 Grb2 Ywhae Bsn Development and Eef2 Ywhaz differentiation Rpgrip1l Atp6v1a Nphp1 Iqcb1 Ezh2 Ywhae Ywhab Pgls Hsd3b7 Hsd3b4 Development and Gnai1 Dlg4 differentiation Development and Gnao1 Dlg4 differentiation Vamp2 App Ywhae Ywhab Development and Psmd3 Rpgrip1l differentiation Psmd4 Hmga2 Development and Thy1 Syp differentiation Atp6v1a App Ywhae Ywhaz Ywhab Hsd3b7 Hsd3b4 Development and Tubb2a Ywhaz differentiation Nphp4 -

An Integrated Approach to Comprehensively Map the Molecular Context of Proteins
bioRxiv preprint doi: https://doi.org/10.1101/264788; this version posted February 13, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Liu et al. An integrated approach to comprehensively map the molecular context of proteins Xiaonan Liu1,2, Kari Salokas1,2, Fitsum Tamene1,2,3 and Markku Varjosalo1,2,3* 1Institute of Biotechnology, University of Helsinki, Helsinki 00014, Finland 2Helsinki Institute of Life Science, University of Helsinki, Helsinki 00014, Finland 3Proteomics Unit, University of Helsinki, Helsinki 00014, Finland *Corresponding author Abstract: Protein-protein interactions underlie almost all cellular functions. The comprehensive mapping of these complex cellular networks of stable and transient associations has been made available by affi nity purifi cation mass spectrometry (AP-MS) and more recently by proximity based labelling methods such as BioID. Due the advancements in both methods and MS instrumentation, an in-depth analysis of the whole human proteome is at grasps. In order to facilitate this, we designed and optimized an integrated approach utilizing MAC-tag combining both AP-MS and BioID in a single construct. We systematically applied this approach to 18 subcellular localization markers and generated a molecular context database, which can be used to defi ne molecular locations for any protein of interest. In addition, we show that by combining the AP-MS and BioID results we can also obtain interaction distances within a complex. Taken together, our combined strategy off ers comprehensive approach for mapping physical and functional protein interactions. Introduction: Majority of proteins do not function in isolation geting the endogenous bait protein, allowing and their interactions with other proteins defi ne purifi cation of the bait protein together with the their cellular functions. -

Supplementary Material Table of Contents
Supplementary M aterial Table of Contents 1 - Online S u pplementary Methods ………………………………………………...… . …2 1.1 Study population……………………………………………………………..2 1.2 Quality control and principal component analysis …………………………..2 1.3 Statistical analyses………………………………………… ………………...3 1.3.1 Disease - specific association testing ……………………………… ..3 1.3.2 Cross - phenotype meta - analysis …………………………………… .3 1.3.3 Model search ……………………………………………………… .4 1.3.4 Novelty of the variants …………………………………………… ..4 1.3.5 Functional Enrichment Analy sis ………………………………… ...4 1.3.6 Drug Target Enrichment Analysis ………………………………… 5 2 - Supplementary Figures………………………………………...………………… . …. 7 3 - Members of the Myositis Genetics Consortium (MYOGEN) ……………………. ..16 4 - Members of the Scleroderma Genetics Consortium ………………… ……………...17 5 - Supplementary References………………………………………………………… . .18 6 - Supplementary Tables………………………………………………………… . ……22 1 Online supplementary m ethods Study population This study was conducted using 12,132 affected subjects and 23 ,260 controls of European des cent population and all of them have been included in previously published GWAS as summarized in Table S1. [1 - 6] Briefly, a total of 3,255 SLE cases and 9,562 ancestry matched controls were included from six countrie s across Europe and North America (Spain, Germany, Netherlands, Italy, UK, and USA). All of the included patients were diagnosed based on the standard American College of Rheumatology (ACR) classification criteria. [7] Previously described GWAS data from 2,363 SSc cases and 5,181 ancestry -

The MUC6/AP2A2 Locus and Its Relevance to Alzheimer’S Disease: a Review
J Neuropathol Exp Neurol Vol. 79, No. 6, June 2020, pp. 568–584 doi: 10.1093/jnen/nlaa024 REVIEW ARTICLE The MUC6/AP2A2 Locus and Its Relevance to Alzheimer’s Disease: A Review Peter T. Nelson, MD, PhD, David W. Fardo, PhD, and Yuriko Katsumata, PhD Abstract INTRODUCTION We recently reported evidence of Alzheimer’s disease (AD)- This review article is about a putative Alzheimer’s dis- linked genetic variation within the mucin 6 (MUC6) gene on chro- ease (AD)-associated genetic polymorphism within the mucin 6 mosome 11p, nearby the adaptor-related protein complex 2 subunit (MUC6) gene, and <4000 base pairs (bp) away from the gene alpha 2 (AP2A2) gene. This locus has interesting features related to that encodes adaptor-related protein complex 2 subunit alpha 2 human genomics and clinical research. MUC6 gene variants have (AP2A2), on human chromosome 11p15.5 (1). The genomic lo- been reported to potentially influence viral—including herpesvi- cus of interest is a poorly annotated variable number of tandem rus—immunity and the gut microbiome. Within the MUC6 gene is a repeat (VNTR) region, largely unreadable by conventional se- unique variable number of tandem repeat (VNTR) region. We dis- quencing methods. Here, we review the topical literature on covered an association between MUC6 VNTR repeat expansion and MUC6, the risk-associated polymorphic VNTR region, and AD pathologic severity, particularly tau proteinopathy. Here, we re- AP2A2. We examine whether it is credible that a novel high- view the relevant literature. The AD-linked VNTR polymorphism impact risk allele could be discovered, given that so many AD- may also influence AP2A2 gene expression. -

Downloaded from the Gene Expression Omnibus (GSE25219) Were Used for the Assessment of Prenatal Gene Expression
UCLA UCLA Electronic Theses and Dissertations Title Investigation of sex-differential genetic risk factors for autism spectrum disorders Permalink https://escholarship.org/uc/item/4cf1h61b Author Werling, Donna Marie Publication Date 2014 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA Los Angeles Investigation of sex-differential genetic risk factors for autism spectrum disorders A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Neuroscience by Donna Marie Werling 2014 © Copyright by Donna Marie Werling 2014 ABSTRACT OF THE DISSERTATION Investigation of sex-differential genetic risk factors for autism spectrum disorders by Donna Marie Werling Doctor of Philosophy in Neuroscience University of California, Los Angeles, 2014 Professor Daniel H. Geschwind, Chair Autism spectrum disorders (ASDs) are pervasive neurodevelopmental disorders that affect more males than females, and the mechanisms responsible for increasing males’ risk or protecting females are not understood. This sex biased prevalence is consistent across time and populations, suggesting that an understanding of the processes driving sex-differential risk would likely be informative of fundamental pathophysiology in ASD. One known component of ASD risk is genetic variation. Thus, here I apply several approaches that leverage current knowledge of ASD genetics to investigate the role and mechanisms of sex-differential biology in ASD risk. First, I evaluate a cohort of families with more than one autistic child for evidence of sex- differential, familial risk variation. Second, I use genetic linkage analysis to identify sex- differential risk loci in families from the same multiplex cohort. -

A SARS-Cov-2 Protein Interaction Map Reveals Targets for Drug Repurposing
Article A SARS-CoV-2 protein interaction map reveals targets for drug repurposing https://doi.org/10.1038/s41586-020-2286-9 A list of authors and affiliations appears at the end of the paper Received: 23 March 2020 Accepted: 22 April 2020 A newly described coronavirus named severe acute respiratory syndrome Published online: 30 April 2020 coronavirus 2 (SARS-CoV-2), which is the causative agent of coronavirus disease 2019 (COVID-19), has infected over 2.3 million people, led to the death of more than Check for updates 160,000 individuals and caused worldwide social and economic disruption1,2. There are no antiviral drugs with proven clinical efcacy for the treatment of COVID-19, nor are there any vaccines that prevent infection with SARS-CoV-2, and eforts to develop drugs and vaccines are hampered by the limited knowledge of the molecular details of how SARS-CoV-2 infects cells. Here we cloned, tagged and expressed 26 of the 29 SARS-CoV-2 proteins in human cells and identifed the human proteins that physically associated with each of the SARS-CoV-2 proteins using afnity-purifcation mass spectrometry, identifying 332 high-confdence protein–protein interactions between SARS-CoV-2 and human proteins. Among these, we identify 66 druggable human proteins or host factors targeted by 69 compounds (of which, 29 drugs are approved by the US Food and Drug Administration, 12 are in clinical trials and 28 are preclinical compounds). We screened a subset of these in multiple viral assays and found two sets of pharmacological agents that displayed antiviral activity: inhibitors of mRNA translation and predicted regulators of the sigma-1 and sigma-2 receptors. -

Involved Genes Carbon Metabolism 7.87E‐09 8.10 IDH3G
Supplemental Table 9: Sulfhydrated Protein Pathway Enrichment in Brain KEGG Pathway (DR) p‐val (adj) ‐LOG10(p‐val adj) Involved Genes Carbon metabolism 7.87E‐09 8.10 IDH3G,DLST,PGAM1,ALDOC,MDH2,OGDH,PFKP,GLUD1,TKT,ACO2,ECHS1,IDH1,CAT,ACAT1,PKM,PCCB,PFKM,GPI,PHGDH,PGK1 Regulation of actin cytoskeleton 0.0102 1.99 PPP1CC,CRKL,PPP1CB,PFN1,VCL,BAIAP2,ARPC5L,PFN2,ARPC1A,PAK1,RDX,EZR,ACTN4,CFL1,CFL2,MAPK1,ARPC4 Synaptic vesicle cycle 0.00658 2.18 AP2A2,NAPA,STX1A,ATP6V1E1,VAMP2,AP2M1,SNAP25,STX1B,ATP6V1A Pentose phosphate pathway 0.024 1.62 ALDOC,PFKP,TKT,PGM1,PFKM,GPI Insulin secretion 0.00276 2.56 ATP1A2,STX1A,VAMP2,CAMK2G,SNAP25,SLC2A1,ATP1A1,ATP1A3,ATP1B2,PRKCB,PCLO Biosynthesis of amino acids 0.000129 3.89 IDH3G,PGAM1,ALDOC,PFKP,TKT,ACO2,IDH1,GLUL,PKM,PFKM,PHGDH,PGK1 Fc gamma R‐mediated phagocytosis 0.0144 1.84 CRKL,ARPC5L,ARPC1A,PAK1,PRKCB,CFL1,CFL2,MAPK1,MARCKS,ARPC4 Glyoxylate and dicarboxylate metabolism 0.0138 1.86 MDH2,ACO2,GLUL,CAT,ACAT1,PCCB Citrate cycle (TCA cycle) 0.0201 1.70 IDH3G,DLST,MDH2,OGDH,ACO2,IDH1 Glycolysis / Gluconeogenesis 0.000216 3.67 PGAM1,ALDOC,PFKP,AKR1A1,PGM1,ALDH2,PKM,PFKM,GPI,PGK1,LDHA Proximal tubule bicarbonate reclamation 0.00209 2.68 ATP1A2,GLUD1,CA2,ATP1A1,ATP1A3,ATP1B2 Oocyte meiosis 0.00228 2.64 PPP1CC,PPP1CB,YWHAB,YWHAH,PPP2CA,YWHAE,CAMK2G,YWHAZ,PPP3CA,SKP1,YWHAG,MAPK1,YWHAQ Gap junction 0.0131 1.88 TUBB6,TUBB5,TUBA4A,TUBB4B,TUBA1C,PRKCB,TUBB2A,TUBB3,TUBB4A,MAPK1 Protein processing in endoplasmic reticulum 0.0171 1.77 PRKCSH,CALR,DNAJB11,HSPA8,PDIA6,HSP90AB1,P4HB,RAD23B,VCP,ERP29,HSPH1,SKP1,FBXO2,GANAB -
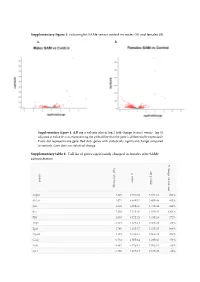
Supplementary Figure 1: Volcano Plot Same Versus Control Im Males (A) and Females (B)
Supplementary figure 1: volcano plot SAMe versus control im males (A) and females (B). A. B. Supplementary figure 1. A,B are a volcano plot of log 2 fold-change (x-axis) versus −log 10 adjusted p-value (y-axis, representing the probability that the gene is differentially expressed). Every dot represents one gene. Red dots: genes with statistically significant change compared to controls. Grey dots: no statistical change. Supplementary table 1: Full list of genes significantly changed in females after SAMe administration % change control-sam log2 fold change ajd p valueajd p value symbol Vegfa 1.829 4.63E-69 3.57E-66 292% Slc2a1 2.072 8.84E-52 3.40E-49 445% Jun 2.608 6.80E-47 1.75E-44 449% Fos 5.288 5.71E-40 1.10E-37 4294% Flt1 2.053 6.72E-21 1.03E-18 272% Thy1 -1.921 1.87E-18 2.40E-16 -66% Egr1 2.745 1.11E-17 1.22E-15 664% Npas4 3.121 3.18E-16 3.06E-14 479% Gfap -1.761 1.50E-14 1.29E-12 -59% Fcrls -1.647 9.75E-13 7.50E-11 -68% Spi1 -1.496 4.01E-12 2.81E-10 -26% Cx3cr1 -1.46 2.29E-11 1.47E-09 -66% Htr1a -1.128 4.77E-10 2.82E-08 -63% C1qb -1.231 1.19E-09 6.55E-08 -31% Cdc40 0.5253 2.32E-09 1.19E-07 59% Calm1 -1.009 3.50E-09 1.68E-07 -54% C1qc -0.9557 5.35E-09 2.42E-07 -24% Stx1a -0.9024 8.78E-09 3.76E-07 -54% Icam1 1.318 2.42E-08 9.79E-07 415% Cd68 -1.109 6.29E-08 0.0000022 -6% Map2k2 -0.4522 6.10E-08 0.0000022 -26% Polr2b 0.3907 5.95E-08 0.0000022 28% Aqp4 -1.849 9.32E-08 3.119E-06 -61% Gusb -0.768 3.88E-07 0.00001194 -28% Polr2l -0.7367 3.78E-07 0.00001194 -43% Cd44 -0.9637 5.08E-07 0.00001506 579% Lypla1 -0.7132 5.48E-07 0.00001561 -3%