First Evidence of Heteroplasmy in Grey Partridge (Perdix
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1471-2148-10-132.Pdf
Shen et al. BMC Evolutionary Biology 2010, 10:132 http://www.biomedcentral.com/1471-2148/10/132 RESEARCH ARTICLE Open Access AResearch mitogenomic article perspective on the ancient, rapid radiation in the Galliformes with an emphasis on the Phasianidae Yong-Yi Shen1,2,3, Lu Liang1,2,3, Yan-Bo Sun1,2,3, Bi-Song Yue4, Xiao-Jun Yang1, Robert W Murphy1,5 and Ya- Ping Zhang*1,2 Abstract Background: The Galliformes is a well-known and widely distributed Order in Aves. The phylogenetic relationships of galliform birds, especially the turkeys, grouse, chickens, quails, and pheasants, have been studied intensively, likely because of their close association with humans. Despite extensive studies, convergent morphological evolution and rapid radiation have resulted in conflicting hypotheses of phylogenetic relationships. Many internal nodes have remained ambiguous. Results: We analyzed the complete mitochondrial (mt) genomes from 34 galliform species, including 14 new mt genomes and 20 published mt genomes, and obtained a single, robust tree. Most of the internal branches were relatively short and the terminal branches long suggesting an ancient, rapid radiation. The Megapodiidae formed the sister group to all other galliforms, followed in sequence by the Cracidae, Odontophoridae and Numididae. The remaining clade included the Phasianidae, Tetraonidae and Meleagrididae. The genus Arborophila was the sister group of the remaining taxa followed by Polyplectron. This was followed by two major clades: ((((Gallus, Bambusicola) Francolinus) (Coturnix, Alectoris)) Pavo) and (((((((Chrysolophus, Phasianus) Lophura) Syrmaticus) Perdix) Pucrasia) (Meleagris, Bonasa)) ((Lophophorus, Tetraophasis) Tragopan))). Conclusions: The traditional hypothesis of monophyletic lineages of pheasants, partridges, peafowls and tragopans was not supported in this study. -

Reproductive Ecology of Tibetan Eared Pheasant Crossoptilon Harmani in Scrub Environment, with Special Reference to the Effect of Food
Ibis (2003), 145, 657–666 Blackwell Publishing Ltd. Reproductive ecology of Tibetan Eared Pheasant Crossoptilon harmani in scrub environment, with special reference to the effect of food XIN LU1* & GUANG-MEI ZHENG2 1Department of Zoology, College of Life Sciences, Wuhan University, Wuhan 430072, China 2Department of Ecology, College of Life Sciences, Beijing Normal University, Beijing 100875, China We studied the nesting ecology of two groups of the endangered Tibetan Eared Pheasants Crossoptilon harmani in scrub environments near Lhasa, Tibet, during 1996 and 1999–2001. One group received artificial food from a nunnery prior to incubation whereas the other fed on natural food. This difference in the birds’ nutritional history allowed us to assess the effects of food on reproduction. Laying occurred between mid-April and early June, with a peak at the end of April or early May. Eggs were laid around noon. Adult females produced one clutch per year. Clutch size averaged 7.4 eggs (4–11). Incubation lasted 24–25 days. We observed a higher nesting success (67.7%) than reported for other eared pheasants. Pro- visioning had no significant effect on the timing of clutch initiation or nesting success, and a weak effect on egg size and clutch size (explaining 8.2% and 9.1% of the observed variation, respectively). These results were attributed to the observation that the unprovisioned birds had not experienced local food shortage before laying, despite spending more time feeding and less time resting than the provisioned birds. Nest-site selection by the pheasants was non-random with respect to environmental variables. Rock-cavities with an entrance aver- aging 0.32 m2 in size and not deeper than 1.5 m were greatly preferred as nest-sites. -

Specimen Record of a Long-Billed Murrelet from Eastern Washington, with Notes on Plumage and Morphometric Differences Between Long-Billed and Marbled Murrelets
SPECIMEN RECORD OF A LONG-BILLED MURRELET FROM EASTERN WASHINGTON, WITH NOTES ON PLUMAGE AND MORPHOMETRIC DIFFERENCES BETWEEN LONG-BILLED AND MARBLED MURRELETS CHRISTOPER W. THOMPSON, WashingtonDepartment of Fish and Wildlife, 16018 Mill Creek Blvd., Mill Creek, Washington98012, and Burke Museum,Box 353100, Universityof Washington,Seattle, Washington 98195 KEVIN J. PULLEN, ConnerMuseum, Washington State University, Pullman, Wash- ington 99164 RICHARD E. JOHNSON, Conner Museumand School of BiologicalSciences, WashingtonState University,Pullman, Washington 99164 ERICB. CUMMINS, WashingtonDepartment of Fishand Wildlife, 600 CapitolWay North, Olympia,Washington 98501 ABSTRACT:On 14 August2001, RobertDice found a Brachyramphusmurrelet approximately12 mileseast of Pomeroyin easternWashington state more than 200 milesfrom the nearestmarine waters. The bird died later that day. It had begun definitiveprebasic body molt, but not flightfeather molt. Necropsy indicated that the birdwas a female,probably in her secondcalendar year. Johnson and Thompson identifiedthe birdas a Long-billedMurrelet, Brachyramphus perdix, on the basisof plumageand measurements;it is the firstspecimen of thisspecies for Washington state. Contrary to many recent publicationsstating that Long-billedand Marbled Murreletshave white and brownunder wing coverts, respectively, we confirmedthat bothspecies typically have white under wing coverts prior to definitiveprebasic molt andbrown under wing coverts after this molt. Absence of anyextensive storm systems in the North Pacificin -

Introduction to Risk Assessments for Methods Used in Wildlife Damage Management
Human Health and Ecological Risk Assessment for the Use of Wildlife Damage Management Methods by USDA-APHIS-Wildlife Services Chapter I Introduction to Risk Assessments for Methods Used in Wildlife Damage Management MAY 2017 Introduction to Risk Assessments for Methods Used in Wildlife Damage Management EXECUTIVE SUMMARY The USDA-APHIS-Wildlife Services (WS) Program completed Risk Assessments for methods used in wildlife damage management in 1992 (USDA 1997). While those Risk Assessments are still valid, for the most part, the WS Program has expanded programs into different areas of wildlife management and wildlife damage management (WDM) such as work on airports, with feral swine and management of other invasive species, disease surveillance and control. Inherently, these programs have expanded the methods being used. Additionally, research has improved the effectiveness and selectiveness of methods being used and made new tools available. Thus, new methods and strategies will be analyzed in these risk assessments to cover the latest methods being used. The risk assements are being completed in Chapters and will be made available on a website, which can be regularly updated. Similar methods are combined into single risk assessments for efficiency; for example Chapter IV contains all foothold traps being used including standard foothold traps, pole traps, and foot cuffs. The Introduction to Risk Assessments is Chapter I and was completed to give an overall summary of the national WS Program. The methods being used and risks to target and nontarget species, people, pets, and the environment, and the issue of humanenss are discussed in this Chapter. From FY11 to FY15, WS had work tasks associated with 53 different methods being used. -

Hybridization & Zoogeographic Patterns in Pheasants
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Paul Johnsgard Collection Papers in the Biological Sciences 1983 Hybridization & Zoogeographic Patterns in Pheasants Paul A. Johnsgard University of Nebraska-Lincoln, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/johnsgard Part of the Ornithology Commons Johnsgard, Paul A., "Hybridization & Zoogeographic Patterns in Pheasants" (1983). Paul Johnsgard Collection. 17. https://digitalcommons.unl.edu/johnsgard/17 This Article is brought to you for free and open access by the Papers in the Biological Sciences at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Paul Johnsgard Collection by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. HYBRIDIZATION & ZOOGEOGRAPHIC PATTERNS IN PHEASANTS PAUL A. JOHNSGARD The purpose of this paper is to infonn members of the W.P.A. of an unusual scientific use of the extent and significance of hybridization among pheasants (tribe Phasianini in the proposed classification of Johnsgard~ 1973). This has occasionally occurred naturally, as for example between such locally sympatric species pairs as the kalij (Lophura leucol11elana) and the silver pheasant (L. nycthelnera), but usually occurs "'accidentally" in captive birds, especially in the absence of conspecific mates. Rarely has it been specifically planned for scientific purposes, such as for obtaining genetic, morphological, or biochemical information on hybrid haemoglobins (Brush. 1967), trans ferins (Crozier, 1967), or immunoelectrophoretic comparisons of blood sera (Sato, Ishi and HiraI, 1967). The literature has been summarized by Gray (1958), Delacour (1977), and Rutgers and Norris (1970). Some of these alleged hybrids, especially those not involving other Galliformes, were inadequately doculnented, and in a few cases such as a supposed hybrid between domestic fowl (Gallus gal/us) and the lyrebird (Menura novaehollandiae) can be discounted. -

Trichostrongylus Cramae N. Sp. (Nematoda), a Parasite of Bob-White Quail (Colinus Virginianus) M.-C
Ann. Parasitol. Hum. Comp., Key-words: Trichostrongylus. Birds. Europe. USA. Trichos- 1993, 68 : n° 1, 43-48. trongylus tenuis. T. cramae n. sp. Lagopus scoticus. Pavo cris- tatus. Perdix perdix. Phasianus colchicus. Colinus virginianus. Mémoire. Mots-clés : Trichostrongylus. Oiseaux. Europe. USA. Trichos trongylus tenuis. T. cramae n. sp. Lagopus scoticus. Pavo cris- tatus. Perdix perdix. Phasianus colchicus. Colinus virginianus. TRICHOSTRONGYLUS CRAMAE N. SP. (NEMATODA), A PARASITE OF BOB-WHITE QUAIL (COLINUS VIRGINIANUS) M.-C. DURETTE-DESSET*, A. G. CHABAUD*, J. MOORE** Summary ---------------------------------------------------------- Cram (1925, 1927) incorrectly identified as T. pergracilis (now the cuticular striation, the relative distances between the second, a synonym of T. tenuis) what was in reality an undescribed spe third and fourth bursal papillae and the configuration of the dorsal cies in Colinus virginianus. ray. Red grouse (Lagopus scoticus), the type host of T. pergra Trichostrongylus cramae n. sp. is proposed for T. pergracilis cilis, was in fact found to be parasitized by T. tenuis, confirming sensu Cram, 1927 nec Cobbold, 1873 from C. virginianus from the synonymy of T. pergracilis and T. tenuis. USA. It differs from T. tenuis (Mehlis in Creplin, 1846) as regards Résumé : Trichostrongylus cramae n. sp. (Nematoda) parasite de Colinus virginianus. Cram (1925, 1927) a identifié par erreur comme étant T. per Il se différencie de T. tenuis (Mehlis in Creplin, 1846) par la gracilis, maintenant considéré comme un synonyme de T. tenuis, striation cuticulaire, les distances relatives entre les papilles bur- ce qui était en réalité une espèce non décrite parasite de Colinus sales 2, 3 et 4, et par la configuration de la côte dorsale. -

Alpha Codes for 2168 Bird Species (And 113 Non-Species Taxa) in Accordance with the 62Nd AOU Supplement (2021), Sorted Taxonomically
Four-letter (English Name) and Six-letter (Scientific Name) Alpha Codes for 2168 Bird Species (and 113 Non-Species Taxa) in accordance with the 62nd AOU Supplement (2021), sorted taxonomically Prepared by Peter Pyle and David F. DeSante The Institute for Bird Populations www.birdpop.org ENGLISH NAME 4-LETTER CODE SCIENTIFIC NAME 6-LETTER CODE Highland Tinamou HITI Nothocercus bonapartei NOTBON Great Tinamou GRTI Tinamus major TINMAJ Little Tinamou LITI Crypturellus soui CRYSOU Thicket Tinamou THTI Crypturellus cinnamomeus CRYCIN Slaty-breasted Tinamou SBTI Crypturellus boucardi CRYBOU Choco Tinamou CHTI Crypturellus kerriae CRYKER White-faced Whistling-Duck WFWD Dendrocygna viduata DENVID Black-bellied Whistling-Duck BBWD Dendrocygna autumnalis DENAUT West Indian Whistling-Duck WIWD Dendrocygna arborea DENARB Fulvous Whistling-Duck FUWD Dendrocygna bicolor DENBIC Emperor Goose EMGO Anser canagicus ANSCAN Snow Goose SNGO Anser caerulescens ANSCAE + Lesser Snow Goose White-morph LSGW Anser caerulescens caerulescens ANSCCA + Lesser Snow Goose Intermediate-morph LSGI Anser caerulescens caerulescens ANSCCA + Lesser Snow Goose Blue-morph LSGB Anser caerulescens caerulescens ANSCCA + Greater Snow Goose White-morph GSGW Anser caerulescens atlantica ANSCAT + Greater Snow Goose Intermediate-morph GSGI Anser caerulescens atlantica ANSCAT + Greater Snow Goose Blue-morph GSGB Anser caerulescens atlantica ANSCAT + Snow X Ross's Goose Hybrid SRGH Anser caerulescens x rossii ANSCAR + Snow/Ross's Goose SRGO Anser caerulescens/rossii ANSCRO Ross's Goose -

Plasma Biochemical Parameters of the Blood of Captive Adult Male and Female Black-Necked Pheasants
ISSN: 2689-4246 DOI: 10.33552/CTCMS.2021.02.000540 Current Trends in Clinical & Medical Sciences Research Article Copyright © All rights are reserved by Slavko Naskov Nikolov Plasma Biochemical Parameters of The Blood of Captive Adult Male and Female Black-Necked Pheasants (Phasianus colchicus), Gray Partridge (Perdix perdix) and Chukar Partridge (Alectoris chukar) in Bulgaria Slavko Naskov Nikolov* and Dian Todorov Kanakov Department Internal non-infectious Diseases, Faculty of Veterinary Medicine, Trakia University, Bulgaria *Corresponding author: Slavko Nikolov, Department of Internal Non-infectious Received Date: January 28, 2021 Diseases, Faculty of Veterinary Medicine; Trakia University, Bulgaria. Published Date: February 05, 2021 Abstract Blood samples were collected from 36 (12 by species; 6 male and 6 female) captive, adult, clinically healthy Black-necked pheasants or Southern Caucasus pheasants (Phasianus colchicus), Gray partridge (Perdix perdix) and Chukar partridge (Alectoris chukar) for plasma biochemical analyses. in concentrations of total bilirubin and Glucose in Gray partridges; and Total protein in Chukar. In female Gray and Chukar partridges, the Total proteinThe investigated values were parameters higher than were in maleTotal ones,protein, but Albumin, in pheasants Total it bilirubin was the opposite. and Glucose. The Significanttrend for Albumin differences and (P<0.05)Total bilirubin among values both insexes the werethree found game birds were inverted, with higher values observed in male than female birds. Keywords: Plasma biochemistry; Blood values; Pheasants; Gray partridge; Chukar; Game birds Introduction chukars [7]. There are studies have reported the values of biochem- The Black-necked pheasants or Southern Caucasus pheas- ical parameters in pheasants [8-9], and the knowledge of plasma ants (Phasianus col. -
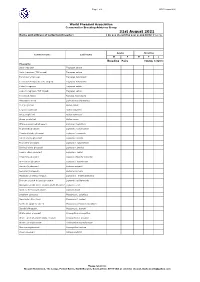
31St August 2021 Name and Address of Collection/Breeder: Do You Closed Ring Your Young Birds? Yes / No
Page 1 of 3 WPA Census 2021 World Pheasant Association Conservation Breeding Advisory Group 31st August 2021 Name and address of collection/breeder: Do you closed ring your young birds? Yes / No Adults Juveniles Common name Latin name M F M F ? Breeding Pairs YOUNG 12 MTH+ Pheasants Satyr tragopan Tragopan satyra Satyr tragopan (TRS ringed) Tragopan satyra Temminck's tragopan Tragopan temminckii Temminck's tragopan (TRT ringed) Tragopan temminckii Cabot's tragopan Tragopan caboti Cabot's tragopan (TRT ringed) Tragopan caboti Koklass pheasant Pucrasia macrolopha Himalayan monal Lophophorus impeyanus Red junglefowl Gallus gallus Ceylon junglefowl Gallus lafayettei Grey junglefowl Gallus sonneratii Green junglefowl Gallus varius White-crested kalij pheasant Lophura l. hamiltoni Nepal Kalij pheasant Lophura l. leucomelana Crawfurd's kalij pheasant Lophura l. crawfurdi Lineated kalij pheasant Lophura l. lineata True silver pheasant Lophura n. nycthemera Berlioz’s silver pheasant Lophura n. berliozi Lewis’s silver pheasant Lophura n. lewisi Edwards's pheasant Lophura edwardsi edwardsi Vietnamese pheasant Lophura e. hatinhensis Swinhoe's pheasant Lophura swinhoii Salvadori's pheasant Lophura inornata Malaysian crestless fireback Lophura e. erythrophthalma Bornean crested fireback pheasant Lophura i. ignita/nobilis Malaysian crestless fireback/Vieillot's Pheasant Lophura i. rufa Siamese fireback pheasant Lophura diardi Southern Cavcasus Phasianus C. colchicus Manchurian Ring Neck Phasianus C. pallasi Northern Japanese Green Phasianus versicolor -

Eastern China
The magnificent Reeves's Pheasant was one of the many specialties seen on this tour (Brendan Ryan). EASTERN CHINA 3 – 27 MAY 2017 LEADER: HANNU JÄNNES Birdquest’s Eastern China tour, an epic 25 day journey across much of eastern China, focusses on an array of rare Chinese endemics and migrants, and this year’s tour once again proved a great success. The focus of the first part of the tour is to achieve good views of rarities like Spoon-billed Sandpiper, the critically endangered Blue-crowned (Courtois’s) Laughingthrush, the superb Cabot’s Tragopan and Elliot’s Pheasant and the ultra-rare Chinese Crested Tern. This was successfully achieved alongside a plethora of other much sought after species including White-faced Plover, Great Knot, stunning Saunders’s Gulls, Reed Parrotbill, eastern migrants, including Pechora Pipit, Japanese Robin, Japanese Paradise, Yellow-rumped, Narcissus and Mugimaki Flycatchers, and forest species like Brown-chested Jungle Flycatcher, White-necklaced Partridge, Silver Pheasant, Buffy and Moustached Laughingthrushes, Short-tailed Parrotbill, Fork-tailed Sunbird and the delightful Pied Falconet. Quite a haul! 1 BirdQuest Tour Report: Eastern China 2017 www.birdquest-tours.com Crested Ibis at Dongzhai Nature Reserve (Brendan Ryan). The second part of the tour, the ‘Northeast Extension’, visited a series of sites for various other Chinese specialities. Beginning in Wuhan, we bagged the amazing Reeves’s Pheasant and Crested Ibis, as well as stunners that included Fairy Pitta and Chestnut-winged Cuckoo. We then moved on to Jiaocheng for the fabulous Brown Eared Pheasants before flying on to Beijing, where the mountains of the nearby Hebei province yielded the endemic Chinese Beautiful Rosefinch, Chinese Nuthatch, Green-backed and Zappey’s Flycatchers and the rare Grey-sided Thrush. -

Blood Plasma Biochemical Parameters of Captive Black-Necked Pheasant
Trakia Journal of Sciences, No 2, pp 142-146, 2021 Copyright © 2021 Trakia University Available online at: http://www.uni-sz.bg ISSN 1313-3551 (online) doi:10.15547/tjs.2021.02.005 Original Contribution BLOOD PLASMA BIOCHEMICAL PARAMETERS OF CAPTIVE BLACK-NECKED PHEASANT (PHASIANUS COLCHICUS), GRAY PARTRIDGE (PERDIX PERDIX) AND CHUKAR PARTRIDGE (ALECTORIS CHUKAR) OF BOTH SEXES IN BULGARIA S. Nikolov*, D. Kanakov Department Internal Non-infectious Diseases, Faculty of Veterinary Medicine, Trakia University, Stara Zagora, Bulgaria ABSTRACT In the study, 36 (n=12 by species, n=6 by sex) blood samples were taken from a captive, adult, clinically healthy, Black-necked pheasants or Southern Caucasus pheasants (Phasianus col. colchicus), Gray partridge (Perdix perdix), and Chukar partridge (Alectoris chukar) for plasma biochemical analyses. The investigated plasma biochemical parameters were Creatinine, Uric acid, Aspartate aminotransferase (AST), and Alanine aminotransferase (ALT). Significant differences (P<0.05) among both sexes were found in the activity of ALT in Black-necked pheasants. Creatinine levels are relatively close in value between the sexes in all three species, slightly higher in males. Uric acid values in male birds are much higher than in female game birds in all three species. AST and ALT activities in male pheasants were higher when compared to the females, while the trend was reversed in sex at Gray partridges. Key words: Plasma biochemistry values, plasma blood analysis, pheasants, Gray partridge, Chukar, game birds. INTRODUCTION partridges and chukars (5, 6). Some studies One of the most common game birds in Europe have researched the biochemical parameters in and Asia are Black-necked pheasants or pheasants (7, 8) but the values of plasma Southern Caucasus pheasants (Phasianus col. -

A Multigene Phylogeny of Galliformes Supports a Single Origin of Erectile Ability in Non-Feathered Facial Traits
J. Avian Biol. 39: 438Á445, 2008 doi: 10.1111/j.2008.0908-8857.04270.x # 2008 The Authors. J. Compilation # 2008 J. Avian Biol. Received 14 May 2007, accepted 5 November 2007 A multigene phylogeny of Galliformes supports a single origin of erectile ability in non-feathered facial traits Rebecca T. Kimball and Edward L. Braun R. T. Kimball (correspondence) and E. L. Braun, Dept. of Zoology, Univ. of Florida, P.O. Box 118525, Gainesville, FL 32611 USA. E-mail: [email protected] Many species in the avian order Galliformes have bare (or ‘‘fleshy’’) regions on their head, ranging from simple featherless regions to specialized structures such as combs or wattles. Sexual selection for these traits has been demonstrated in several species within the largest galliform family, the Phasianidae, though it has also been suggested that such traits are important in heat loss. These fleshy traits exhibit substantial variation in shape, color, location and use in displays, raising the question of whether these traits are homologous. To examine the evolution of fleshy traits, we estimated the phylogeny of galliforms using sequences from four nuclear loci and two mitochondrial regions. The resulting phylogeny suggests multiple gains and/or losses of fleshy traits. However, it also indicated that the ability to erect rapidly the fleshy traits is restricted to a single, well-supported lineage that includes species such as the wild turkey Meleagris gallopavo and ring-necked pheasant Phasianus colchicus. The most parsimonious interpretation of this result is a single evolution of the physiological mechanisms that underlie trait erection despite the variation in color, location, and structure of fleshy traits that suggest other aspects of the traits may not be homologous.