Unravelling the Exposome: Conclu- Sions and Thoughts for the Future
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Exposure Science and Environmental Epidemiology (2013) 23, 455–456 & 2013 Nature America, Inc
Journal of Exposure Science and Environmental Epidemiology (2013) 23, 455–456 & 2013 Nature America, Inc. All rights reserved 1559-0631/13 www.nature.com/jes EDITORIAL Exposure science: A need to focus on conducting scientific studies, rather than debating its concepts Journal of Exposure Science and Environmental Epidemiology (2013) establish baseline distributions of many chemicals that can be 23, 455–456; doi:10.1038/jes.2013.43 found in blood and urine; and biomarkers referred to as Biological Exposure Indices (BEIs) have been increasingly used in occupa- tional settings to establish worker exposures since the 1980s. These uses predate the idea of the ‘‘exposome’’ and are important measurements in the exposure science ‘‘toolbox’’. The energy Over the past 25 þ years, Exposure Science has provided a wealth associated with the promotion of the exposome concept, how- of credible experimental, field and theoretical-based research, ever, has been the interpreted, by some, to promote internal including the how, where and when elevated exposures occur, markers of exposure as the best, or as the only, way to employ the and ultimately how to mitigate or prevent contact with chemical principles of exposure science. To date, many internal markers of contaminants and other stressors. It is therefore puzzling that, exposure have only been able to define that ‘‘you have been over the past 7 years, many Exposure Scientists have not taken exposed’’ and not how, when, where, or to how much. advantage of our successes, but instead have focused on the Some internal markers have been more successful in improving adequacy of its Scientific Concepts and Terminology. -

APT Curriculum Vitea
CURRICULUM VITAE Notarization. I have read the following and certify that this curriculum vitae is a current and accurate statement of my professional record. Signature Date April 9, 2021 I. PERSONAL INFORMATION I.A. UID: 114503213 Roberts, Jennifer, Denise Department of Kinesiology University of Maryland School of Public Health 2136 SPH Building #255 4200 Valley Drive College Park, Maryland 20742 Office Phone: 301.405.7748 Email: [email protected] Twitter: @ActiveRoberts Faculty Profile Website: http://sph.umd.edu/people/jennifer-d-roberts PHOEBE Lab Website: www.sph.umd.edu/phoebelab Professional Website: http://www.jenniferdeniseroberts.com I.B. Academic Appointments at UMD 2015 – Present Assistant Professor, Department of Kinesiology, University of Maryland School of Public Health (UMD-SPH), College Park, MD 2015 – Present Director, Public Health Outcomes and Effects of the Built Environment ( PHOEBE) Laboratory, University of Maryland, College Park, MD 2015 – Present Faculty Affiliate, Maryland Population Research Center, University of Maryland, College Park, MD 2017 – Present Faculty Affiliate, Consortium on Race, Gender & Ethnicity, University of Maryland, College Park, MD 2018 – Present Faculty Affiliate, Maryland Transportation Institute, University of Maryland, College Park, MD 2019 – Present Associate Member, NIDDK-Funded Mid-Atlantic Nutrition Obesity Research Center, University of Maryland, School of Medicine, Baltimore, MD I.C. Other Employment 1 2011 – 2015 Assistant Professor, Uniformed Services University (USUHS), F. Edward -

Guidelines for Human Exposure Assessment Risk Assessment Forum
EPA/100/B-19/001 October 2019 www.epa.gov/risk Guidelines for Human Exposure Assessment Risk Assessment Forum EPA/100/B-19/001 October 2019 Guidelines for Human Exposure Assessment Risk Assessment Forum U.S. Environmental Protection Agency DISCLAIMER This document has been reviewed in accordance with U.S. Environmental Protection Agency (EPA) policy. Mention of trade names or commercial products does not constitute endorsement or recommendation for use. Preferred citation: U.S. EPA (U.S. Environmental Protection Agency). (2019). Guidelines for Human Exposure Assessment. (EPA/100/B-19/001). Washington, D.C.: Risk Assessment Forum, U.S. EPA. Page | ii TABLE OF CONTENTS DISCLAIMER ..................................................................................................................................... ii LIST OF TABLES.............................................................................................................................. vii LIST OF FIGURES ........................................................................................................................... viii LIST OF BOXES ................................................................................................................................ ix ABBREVIATIONS AND ACRONYMS .............................................................................................. x PREFACE ........................................................................................................................................... xi AUTHORS, CONTRIBUTORS AND -

Gloria Post, Ph.D., DABT
Updated 9/26/19 Curriculum Vitae Gloria B. Post, Ph.D., D.A.B.T. Office: Division of Science and Research New Jersey Department of Environmental Protection Mail Code 428-01 PO Box 420 Trenton, NJ 08625-0420 Telephone: (609) 292-8497 Fax: (609) 292-7340 Email: [email protected] Home: 5 Teal Drive Langhorne, PA 19047 Education: Ph.D., Pharmacology. Thomas Jefferson University, Philadelphia, PA, 1982. Doctoral thesis topic: Effects of enzyme induction on microsomal benzene metabolism. Advisor: Dr. Robert Snyder A.B. with Honors. Biochemical Sciences, Princeton University, Princeton, NJ, 1977. Senior thesis topic: On the effect of multiple copies of the oligopeptide permease gene in E. coli. Advisor: Dr. Charles Gilvarg Certification: Diplomate of the American Board of Toxicology (since 1990, with recertifications). Professional Experience: Research Scientist, Division of Science and Research, New Jersey Department of Environmental Protection, Trenton, NJ (1986-present). General Responsibilities Responsible for development of human health criteria, standards, and guidance for drinking water, ground water, surface water, soil, and fish consumption, including risk assessment approaches, toxicological basis, and exposure assumptions. Member of the New Jersey Drinking Water Quality Institute (DWQI), a legislatively established advisory body that recommends drinking water standards to NJDEP, since 2006. Provide toxicology and risk assessment support for Health Effects Subcommittee of the DWQI (since 1986). Primary author or co-author of numerous DWQI Health-based Maximum Contaminant Level Support documents posted at http://www.nj.gov/dep/watersupply/g_boards_dwqi.html Provide technical support to NJDEP programs on health effects, risk assessment, and other topics related to per- and polyfluoroalkyl substance (PFAS) in drinking water and other environmental media. -

Completing the Link Between Exposure Science and Toxicology for Improved Environmental Health Decision Making
Feature pubs.acs.org/est Completing the Link between Exposure Science and Toxicology for Improved Environmental Health Decision Making: The Aggregate Exposure Pathway Framework † ‡ ○ ∥ ○ ⊥ # Justin G. Teeguarden,*, , , Yu-Mei Tan, , Stephen W. Edwards, Jeremy A. Leonard, ‡ ‡ † § ‡ ‡ Kim A. Anderson, Richard A. Corley, , Molly L. Kile, Staci M. Simonich, David Stone, ‡ ‡ † ‡ ∇ ‡ Robert L. Tanguay, Katrina M. Waters, , Stacey L. Harper, , and David E. Williams † Health Effects and Exposure Science, Pacific Northwest National Laboratory, Richland, Washington 99352, United States ‡ Department of Environmental and Molecular Toxicology, Oregon State University, Corvallis, Oregon 93771, United States § School of Biological and Population Health Sciences, Oregon State University, Corvallis, Oregon 93771, United States ∥ National Exposure Research Laboratory, U.S. Environmental Protection Agency, Durham, North Carolina 27709, United States ⊥ National Health and Environmental Effects Research Laboratory, U.S. Environmental Protection Agency, Durham, North Carolina 27709, United States # Oak Ridge Institute for Science and Education, Oak Ridge, Tennessee 37831, United States ∇ School of Chemical, Biological and Environmental Engineering, Oregon State University, Corvallis, Oregon 97331, United States purpose of protecting ecologic and public health.1 Historically, exposure assessment has played a complementary role with the fields of epidemiology and toxicology, helping identify and mitigate health impacts of environmental exposures, of which lead and radon serve as good examples.1 Recognizing the historical value of exposure science and recent demands to meet the growing need to conduct more comprehensive exposure assessment (thousands of stressors), fi more quickly and more accurately, a committee of the National Driven by major scienti c advances in analytical methods, Academy of Sciences (NAS) recently called for an extensive biomonitoring, computation, and a newly articulated vision for 1 expansion of human and ecological exposure assessment. -

Principles of Characterizing and Applying Human Exposure Models
1 2 3 4 5 6 7 Harmonization Project Document No. X 8 9 10 PRINCIPLES OF CHARACTERIZING AND APPLYING 11 HUMAN EXPOSURE MODELS 12 13 14 15 DRAFT 16 17 18 Prepared by the 19 20 Exposure Modelling Subcommittee of the IPCS Harmonization Project Exposure Assessment 21 Working Group 22 23 International Programme on Chemical Safety 24 World Health Organization 25 Geneva 26 27 28 29 November 2004 30 Harmonization Project Document No. X 1 TABLE OF CONTENTS 2 3 FOREWORD............................................................................................................................4 4 5 LIST OF MEMBERS OF THE EXPOSURE MODELLING SUBCOMMITTEE OF 6 THE IPCS HARMONIZATION PROJECT EXPOSURE ASSESSMENT WORKING 7 GROUP .....................................................................................................................................6 8 9 LIST OF CONTRIBUTORS ..................................................................................................6 10 11 LIST OF ACRONYMS AND ABBREVIATIONS ...............................................................8 12 13 1. INTRODUCTION..........................................................................................................10 14 1.1 Background and rationale ............................................................................................10 15 1.2 Goals of this work........................................................................................................10 16 1.3 Definitions and terminology ........................................................................................11 -

Requirements for the MS in Exposure Science (For Students Entering Autumn 2020 Or Later)
Requirements for the MS in Exposure Science (for students entering autumn 2020 or later) DEOHS Core Requirements 1. ENV H 580: Students are required to complete 3 quarters of this 1-credit course Choose one: for a total of 3 credits. BIOST 511 Medical Biometry I [A] OR (4) 2. ENV H 583 requires that students take 2 credits of either ENV H 700 (Thesis BIOST 517 Applied Biostatistics I [A] (4) Preparation) or 600 (Independent Study) concurrently. If ENV H 700 is taken as EPI 511 Introduction to Epidemiology [A] 4 part of this requirement, those 2 credits can count towards the minimum 9 ENV H 501 Foundations of Env. & Occ. Health [A] 4 credit ENV H 700 requirement. ENV H 502 Assessing & Managing Risks from Human Exposure 4 3. Students select ENV H electives in consultation with their faculty advisor. Non- to Env. Contaminants [W] ENV H electives will be approved on a case-by-case basis. ENV H 580 Env. & Occupational Health Seminar [A,W,Sp] 1+1+1=31 ENV H 583 Thesis Research Proposal Preparation [Sp] 1(+2)2 Minimum Credit Subtotal 20 Option Specific Requirements Choose one: BIOST 512 Medical Biometry II [W] OR (4) BIOST 518 Applied Biostatistics II [W] (4) ENV H 503 Adverse Health Effects of Env. & Occ. Toxicants [Sp] 4 ENV H 553 Env. Exposure Monitoring Methods [W] 4 ENV H 557 Exposure Controls [W] 3 Minimum Credit Subtotal 15 Culminating Experience ENV H 700 Master’s Thesis [E] 9 [A] = Typically offered in autumn quarter Electives [W] = Typically offered in winter quarter Additional elective credits as needed to reach total TBD Var. -

Influence of the Urban Exposome on Birth Weight Mark J
Influence of the Urban Exposome on Birth Weight Mark J. Nieuwenhuijsen, Lydiane Agier, Xavier Basagana, Jose Urquiza, Ibon Tamayo-Uria, Lise Giorgis-Allemand, Oliver Robinson, Valérie Siroux, Léa Maitre, Montserrat de Castro, et al. To cite this version: Mark J. Nieuwenhuijsen, Lydiane Agier, Xavier Basagana, Jose Urquiza, Ibon Tamayo-Uria, et al.. Influence of the Urban Exposome on Birth Weight. Environmental Health Perspectives, National Institute of Environmental Health Sciences, 2019, 127 (4), 14 p. 10.1289/EHP3971. hal-02492187v2 HAL Id: hal-02492187 https://hal.archives-ouvertes.fr/hal-02492187v2 Submitted on 27 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. A Section 508–conformant HTML version of this article Research is available at https://doi.org/10.1289/EHP3971. Influence of the Urban Exposome on Birth Weight Mark J. Nieuwenhuijsen,1,2,3* Lydiane Agier,4* Xavier Basagaña,1,2,3 Jose Urquiza,1,2,3 Ibon Tamayo-Uria,5 Lise Giorgis-Allemand,4 Oliver Robinson,6 Valérie Siroux,4 Léa Maitre,1,2,3 Montserrat de Castro,1,2,3 Antonia Valentin,1,2,3 David Donaire,1,2,3 Payam Dadvand,1,2,3 Gunn Marit Aasvang,7 Norun Hjertager Krog,7 Per E. -
Thought Leadership Published 4Q 2019
THOUGHT LEADERSHIP PUBLISHED 4Q 2019 The Fundamental Role of Exposure Science in Human Health Risk Assessment November 26, 2019 Exposure science focuses on characterizing the distribution and determinants of human exposure to chemical, physical, or biological substances potentially affecting health. Exposure distribution is influenced by the intensity, frequency, and duration of exposures; as such, these are typically the parameters that define exposure characterizations. Exposure determinants are factors that influence exposures, either directly or indirectly, such as lifestyle choices, behavioral patterns, and demographic characteristics. Exposure science plays a fundamental role in human including those that may be found in consumer products, health risk assessment because without exposure, “unless the exposure is low enough to pose no significant there is no health risk. Exposures can be characterized risk of cancer or is significantly below levels observed to qualitatively or quantitatively, and the selected approach cause birth defects or other reproductive harm” (OEHHA is often multifactorial with consideration given to available 2019). In other words, exposure to a listed chemical in a methodology, budget constraints, and participant burden, consumer product at a certain level triggers the regulatory among other factors. Depending on the approach, the requirement for a warning, not the mere presence of the exposure characterization may be associated with some listed chemical in the product. level of error, which, in turn, will influence the validity of the health risk assessment. I describe below two examples that demonstrate the critical role of exposure science in evaluating health Exposure science is essential to epidemiologic study risk in epidemiology and in consumer product safety design. -

2016 Meeting Program
International Society of Exposure Science PROGRAM 2016 Annual Meeting BOOK ONE-PAGE MEETING-AT-A-GLANCE Welcome Messages - Lesa and Yuri _______________________________________________________________________________ 4 TABLE OF Time Sunday Monday Tuesday Wednesday Thursday Time 9 October 10 October 11 October 12 October 13 October - Tim Buckley _________________________________________________________________________________ 5 Meeting-at-a-Glance CONTENTS 07.00 - 08.00 Student/ 07.00 - 08.00 New Researcher - Sunday, October 9 _______________________________________________________________________ 6 Breakfast Workhop - Monday, October 10 ____________________________________________________________________ 6 08:00 - 08:30 08:00 - 08:30 PLENARY PLENARY PLENARY - Tuesday, October 11 ____________________________________________________________________ 7 08:30 - 09:00 90 m Oral 08:30 - 09:00 - Wednesday, October 12 ______________________________________________________________ 9 - Thursday, October 13 ________________________________________________________________ 10 09:00 - 09:30 90 m Oral 90 m Oral 90 m Oral 09:00 - 09:30 ISES Organization ______________________________________________________________________________________ 11 09:30 - 10:00 09:30 - 10:00 General Information _________________________________________________________________________________ 17 Optional ISES Jaarbeurs map ___________________________________________________________________________________________ 19 Work- Board 10:00 - 10:30 POSTER VIEWING 10:00 - 10:30 shops -
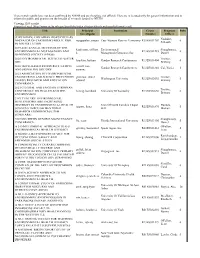
These Search Results Have Not Been Confirmed by NIEHS and Are Therefore, Not Official
These search results have not been confirmed by NIEHS and are therefore, not official. They are to be used only for general information and to inform the public and grantees on the breadth of research funded by NIEHS. Viewing 1289 results Export to Excel (http://www.niehs.nih.gov//portfolio/index.cfm/portfolio/searchresults/format/excel) Title Principal Institution Grant Program Pubs Investigator Number Officer (CIRCADIAN) CIRCADIAN DISRUPTION AS Nadadur, MEDIATOR OF CARDIOMETABOLIC RISK rajagopalan, sanjay Case Western Reserve University R35ES031702 0 Srikanth IN AIR POLLUTION 2019-2021 ANNUAL MEETINGS OF THE kaufmann, william Environmental Shaughnessy, ENVIRONMENTAL MUTAGENESIS AND R13ES031435 0 k. Mutagenesis/Genomics Soc Daniel GENOMICS SOCIETY (EMGS) 2020 ENVIRONMENTAL SCIENCES: WATER Trottier, hsu-kim, heileen Gordon Research Conferences R13ES031404 0 GRC Brittany 2020 THIOL-BASED REDOX REGULATION carroll, kate Gordon Research Conferences R13ES031840 Cui, Yuxia 0 AND SIGNALING GRC/GRS suzanne 2021 ASSOCIATION OF ENVIRONMENTAL ENGINEERING AND SCIENCE PROFESSORS giammar, daniel Trottier, Washington University R13ES033092 0 (AEESP) RESEARCH AND EDUCATION edward Brittany CONFERENCE 2021 CENTRAL AND EASTERN EUROPEAN Trottier, CONFERENCE ON HEALTH AND THE hennig, bernhard University Of Kentucky R13ES032658 0 Brittany ENVIRONMENT 21ST CENTURY ENVIRONMENTAL HEALTH SCHOLARS: INCREASING DIVERSITY IN ENVIRONMENTAL HEALTH Univ Of North Carolina Chapel Humble, jaspers, ilona R25ES031870 0 SCIENCES THROUGH MENTORED Hill Michael RESEARCH EXPERIENCES -
Using 21St Century Science for Risk-Related Evaluations
“Using 21st Century Science for Risk-related evaluations: An Overview of the NAS Risk21 Report” Committee on Incorporating 21st Century Science into Risk-Based Evaluations Board on Environmental Studies and Toxicology Division on Earth and Life Studies Tox21 and ES21 Reports Toxicity Testing in the 21st Century: A Vision and a Strategy was published in 2007 and envisioned a future in which toxicology relied primarily on high-throughput in vitro assays and computational models based on human biology to evaluate potential adverse effects of chemical exposure. Exposure Science in the 21st Century: A Vision and a Strategy was published in 2012 and provided a vision that was hoped to inspire transformational changes in the breadth and depth of exposure assessment that would improve integration with and responsiveness to toxicology and epidemiology. Statement of Task Overall, the committee was asked to provide recommendations on integrating new scientific approaches into risk-based evaluations. The committee was asked to consider the following: The advances that have occurred since publication of Tox21 and ES21 reports. Whether a new paradigm is needed for data validation. How uncertainty evaluations might need to change. What approaches are needed for communicating new science. Barriers and obstacles impeding integration of new science. Sponsors US Environmental Protection Agency US Food and Drug Administration National Institute of Environmental Health Sciences National Center for Advancing Translational Sciences Committee JONATHAN