Earthworm, Perionyx Excavatus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Download Article (PDF)
Rec. zool. Surv. India: l08(Part-3) : 21-25,2008 EARTHWORMS OF NORTH 24 PARGANAS, WEST BENGAL PROBIR K. BANDYOPADHYAY*, C. K. MANDAL** AND AMLAN K. MITRA* * Parasitology Laboratory, Department of Zoology, University of Kalyani, Kalyani-74I 235, West Bengal, India INTRODUCTION Soil animals may play a range of roles in vineyards. Decomposers (some of which are opportunistic herbivores) are important in nutrient dynamics, because by reducing organic matter to its constituents, they liberate nutrients usable by grapevines. Earthworms are only part of the complex of organisms termed "decomposers" in agroecology. As noted by Charles Darwin in his 1882 classic, The Fonnation of Vegetable Mould Through the Action of Earthworms with Observations on Their Habits (Werner, 1990), earthworms process huge quantities of plant litter and help to convert it into rich topsoil, liberating nutrients for renewed plant growth. More recent studies show that earthworms can help to reduce soil compaction, improving permeability and aeration. Earthworms do this through burrowing activities, ingestion of soil along with plant debris, and subsequent excretion of casts. Upon drying, these casts form water-stable soil aggregates. These aggregates are clumps of soil particles bound together by organic compounds, and their presence helps to improve soil structure, retain nutrients that might otherwise be leached, and reduce the threat of erosion (Lee, 1985). Earthworms are increasingly recognized as indicators of agro-ecosystem health and as important tools for ensuring soil improvement and efficient nutrient cycling. In India, due to continuous biodiversity surveys of earthworms number of new species IS increasing day by day, although in comparison to more than 3000 global species (Stephenson, 1923), the number of Indian species is far less (only 390). -

Biological and Physiological Responses of Perionyx Excavatus to Abamectin
Environmental Science and Pollution Research https://doi.org/10.1007/s11356-019-06013-0 RESEARCH ARTICLE Biological and physiological responses of Perionyx excavatus to abamectin Beewah Ng1 & Ratmanee Chanabun2 & Somsak Panha1 Received: 31 January 2019 /Accepted: 22 July 2019 # Springer-Verlag GmbH Germany, part of Springer Nature 2019 Abstract Biological and behavioral responses of the tropical earthworm Perionyx excavatus towards different concentrations of abamectin were evaluated. Abamectin significantly reduced the biomass and reproduction (cocoon production) of P. excavatus as well as inducing histopathological alterations in the cuticle. Biomass loss was recorded in P. excavatus exposed to abamectin at a concentration as low as 0.1 mg active ingredient (a.i.) kg−1, while atrophy, another physiological response, was observed at an abamectin concentration of 0.21 μgcm−2 in a filter paper test. Cocoon production was significantly reduced in the presence of abamectin, and no cocoons were produced at doses of 20 mg a.i. kg−1 or higher, while abamectin at 50 mg a.i. kg−1 induced extreme pathology, characterized by the loss of the integrity of the whole body wall and intestine of P. excavatus. Histopathological alterations can be used as a biomarker to evaluate the toxicological impact of exposure to abamectin. Keywords Abamectin . Perionyx excavatus . Physiological morphology . Cocoon production . Histopathology Introduction tomato russet mite (Aculops lycopersici), armyworms (Spodoptera sp.), and tomato pinworms (Keiferia Agriculture remains an important sector of Malaysia’secono- lycopersicella). The first record of P. xylostella in Malaysia my, contributing 12% to the national gross domestic product was reported in 1925 and it became a serious pest of crucifers (GDP) and providing employment for 16% of the population. -

A Case Study of the Exotic Peregrine Earthworm Morphospecies Pontoscolex Corethrurus Shabnam Taheri, Céline Pelosi, Lise Dupont
Harmful or useful? A case study of the exotic peregrine earthworm morphospecies Pontoscolex corethrurus Shabnam Taheri, Céline Pelosi, Lise Dupont To cite this version: Shabnam Taheri, Céline Pelosi, Lise Dupont. Harmful or useful? A case study of the exotic peregrine earthworm morphospecies Pontoscolex corethrurus. Soil Biology and Biochemistry, Elsevier, 2018, 116, pp.277-289. 10.1016/j.soilbio.2017.10.030. hal-01628085 HAL Id: hal-01628085 https://hal.archives-ouvertes.fr/hal-01628085 Submitted on 5 Jan 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Harmful or useful? A case study of the exotic peregrine earthworm MARK morphospecies Pontoscolex corethrurus ∗ ∗∗ S. Taheria, , C. Pelosib, L. Duponta, a Université Paris Est Créteil, Université Pierre et Marie Curie, CNRS, INRA, IRD, Université Paris-Diderot, Institut d’écologie et des Sciences de l'environnement de Paris (iEES-Paris), Créteil, France b UMR ECOSYS, INRA, AgroParisTech, Université Paris-Saclay, 78026 Versailles, France ABSTRACT Exotic peregrine earthworms are often considered to cause environmental harm and to have a negative impact on native species, but, as ecosystem engineers, they enhance soil physical properties. Pontoscolex corethrurus is by far the most studied morphospecies and is also the most widespread in tropical areas. -

Species Diversity of Terrestrial Earthworms in Different
SPECIES DIVERSITY OF TERRESTRIAL EARTHWORMS IN KHAO YAI NATIONAL PARK Prasuk Kosavititkul A Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in Environmental Biology Suranaree University of Technology Academic Year 2005 ISBN 974-533-516-9 ความหลากหลายของชนิดไสเดือนดินในเขตอุทยานแหงชาติเขาใหญ นายประสุข โฆษวิฑิตกุล วิทยานิพนธน เปี้ นสวนหนงของการศึ่ ึกษาตามหลักสูตรปริญญาวิทยาศาสตรดุษฎีบณฑั ิต สาขาวิชาชีววทยาสิ ิ่งแวดลอม มหาวิทยาลัยเทคโนโลยีสุรนารี ปการศึกษา 2548 ISBN 974-533-516-9 ACKNOWLEDGEMENTS I am most grateful to thank Asst. Prof. Dr. Panee Wannitikul, my advisor, for her generous help, encouragement and guidance throughout this thesis from the beginning. Her criticism, improvement, and proper of manuscript have made this thesis in correct form. I sincerely thank to Assoc. Prof. Dr. Korakod Indrapichate, Assoc. Prof. Dr. Somsak Panha, Dr. Nathawut Thanee and Dr. Pongthep Suwanwaree for their valuable advice and guidance in this thesis. I am sincerely grateful to Assoc. Prof. Dr. Sam James, Natural History Museum and Biodiversity Research Center, Kansas University for his helpful and kind assistance in confirming and identifying earthworms. Thanks also to the Center for Scientific and Technological Equipment, Suranaree University of Technology for the laboratory facilities and scientific instruments. Special thank is due to the Khao Yai National Park for permitting me to work in the park. I would like to express my special thank to Naresuan University for the scholarship supporting my study. Special gratitude is expressed to my parents, my sisters, my family, my friends, my seniors, my juniors, and other people who give me a supported power whenever I lose my own power. Prasuk Kosavititkul CONTENTS Page ABSTRACT IN THAI…………………………………………………………… I ABSTRACT IN ENGLISH……………………………………………………… II ACKNOWLEDGEMENTS…………………………………………………….. -

Biodiversity of Compost Mesofauna and Its Potential As an Indicator of the Composting Process Status
® Dynamic Soil, Dynamic Plant ©2011 Global Science Books Biodiversity of Compost Mesofauna and its Potential as an Indicator of the Composting Process Status Hanne Steel* • Wim Bert Nematology Unit, Department of Biology, Ghent University, K.L. Ledeganckstraat 35, 9000 Ghent, Belgium Corresponding author : * [email protected] ABSTRACT One of the key issues in compost research is to assess the quality and maturity of the compost. Biological parameters, especially based on mesofauna, have multiple advantages for monitoring a given system. The mesofauna of compost includes Isopoda, Myriapoda, Acari, Collembola, Oligochaeta, Tardigrada, Hexapoda, and Nematoda. This wide spectrum of organisms forms a complex and rapidly changing community. Up to the present, none of the dynamics, in relation to the composting process, of these taxa have been thoroughly investigated. However, from the mesofauna, only nematodes possess the necessary attributes to be potentially useful ecological indicators in compost. They occur in any compost pile that is investigated and in virtually all stages of the compost process. Compost nematodes can be placed into at least three functional or trophic groups. They occupy key positions in the compost food web and have a rapid respond to changes in the microbial activity that is translated in the proportion of functional (feeding) groups within a nematode community. Further- more, there is a clear relationship between structure and function: the feeding behavior is easily deduced from the structure of the mouth cavity and pharynx. Thus, evaluation and interpretation of the abundance and function of nematode faunal assemblages or community structures offers an in situ assessment of the compost process. -

F:\Zoos'p~1\2004\March2~1
CASE REPORT ZOOS' PRINT JOURNAL 19(3): 1394-1400 ILLUSTRATIONS OF EARTHWORMS OCCURING IN AND AROUND CHENNAI, INDIA V.I. Ramzan Begum 1 and Sultan Ahmed Ismail 2 1 Institute of Research in Soil Biology and Biotechnology, The New College, Chennai, Tamil Nadu 600014, India. 2 Corresponding author: Ecoscience Research Foundation, Plot 98, Baaz Nagar, 3/621 East Coast Road, Palavakkam, Chennai, Tamil Nadu 600041, India. 2 Email: [email protected] Abstract made to keep the description simple. Earthworms were collected from Chennai and its environs for taxonomic listing and to prepare a brief guide for Earthworms occur all over the world, but only rarely in deserts, identification by budding vermiculturists. Earthworm areas under constant snow and ice, mountain ranges and areas species were identified using existing taxonomic keys. almost entirely lacking in soil and vegetation (Edwards & Bohlen, Drawida scandens, Lampito mauritii, Perionyx 1996). excavatus, Perionyx sansibaricus, Octochaetona barnesi, Octochaetona pattoni, Octochaetona serrata, Earthworms belong to the Order Oligochaeta of the Phylum Octochaetona thurstoni, Dichogaster bolaui and Annelida. Oligochaetes are bilaterally symmetrical coelomate Eudrilus eugeniae have been recorded. D. bolaui was invertebrates with internal and external metameric segmentation the smallest while O. thurstoni was the largest species in throughout the body. Oligochaetes are often divided into two size observed. Fully mature adults of the common species convenient groups Microdrili and Megadrili. During the last collected over a period of time from sites in and around decade and half (Ismail, 1997) much taxonomic work on Indian Chennai are illustrated. Among the species, Eudrilus earthworms has been carried out by Julka (1975, 1976a,b, 1977, eugeniae does not occur naturally and are procured from 1978, 1979, 1981, 1983, 1993), Jamieson (1977) and Easton (1982). -
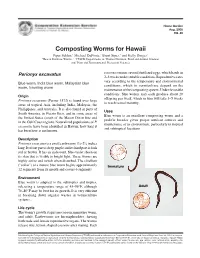
Composting Worms for Hawaii
Home Garden Aug. 2005 HG-46 Composting Worms for Hawaii Piper Selden,1 Michael DuPonte,2 Brent Sipes,3 and Kelly Dinges2 1Hawaii Rainbow Worms, 2, 3CTAHR Departments of 2Human Nutrition, Food and Animal Sciences and 3Plant and Environmental Protection Sciences Perionyx excavatus cocoon contains several fertilized eggs, which hatch in 2–3 weeks under suitable conditions. Reproductive rates Blue worm, India blue worm, Malaysian blue vary according to the temperature and environmental worm, traveling worm conditions, which in vermiculture depend on the maintenance of the composting system. Under favorable Origin conditions, blue worms may each produce about 20 offspring per week, which in turn will take 3–5 weeks Perionyx excavatus (Perrier 1872) is found over large to reach sexual maturity. areas of tropical Asia, including India, Malaysia, the Phillippines, and Australia. It is also found in parts of Uses South America, in Puerto Rico, and in some areas of Blue worm is an excellent composting worm and a the United States (south of the Mason Dixon line and prolific breeder given proper nutrient sources and in the Gulf Coast region). Naturalized populations of P. maintenance of its environment, particularly in tropical excavatus have been identified in Hawaii; how long it and subtropical locations. has been here is not known. Description 1 3 Perionyx excavatus is a small earthworm 1 ⁄4–2 ⁄4 inches long. Its front part is deep purple and its hind part is dark red or brown. It has an iridescent, blue-violet sheen on its skin that is visible in bright light. These worms are highly active and twitch when disturbed. -

Phylogenetic and Phenetic Systematics of The
195 PHYLOGENETICAND PHENETICSYSTEMATICS OF THE OPISTHOP0ROUSOLIGOCHAETA (ANNELIDA: CLITELLATA) B.G.M. Janieson Departnent of Zoology University of Queensland Brisbane, Australia 4067 Received September20, L977 ABSTMCT: The nethods of Hennig for deducing phylogeny have been adapted for computer and a phylogran has been constructed together with a stereo- phylogran utilizing principle coordinates, for alL farnilies of opisthopor- ous oligochaetes, that is, the Oligochaeta with the exception of the Lunbriculida and Tubificina. A phenogran based on the sane attributes conpares unfavourably with the phyLogralnsin establishing an acceptable classification., Hennigrs principle that sister-groups be given equal rank has not been followed for every group to avoid elevation of the more plesionorph, basal cLades to inacceptabl.y high ranks, the 0ligochaeta being retained as a Subclass of the class Clitellata. Three orders are recognized: the LumbricuLida and Tubificida, which were not conputed and the affinities of which require further investigation, and the Haplotaxida, computed. The Order Haplotaxida corresponds preciseLy with the Suborder Opisthopora of Michaelsen or the Sectio Diplotesticulata of Yanaguchi. Four suborders of the Haplotaxida are recognized, the Haplotaxina, Alluroidina, Monil.igastrina and Lunbricina. The Haplotaxina and Monili- gastrina retain each a single superfanily and fanily. The Alluroidina contains the superfamiJ.y All"uroidoidea with the fanilies Alluroididae and Syngenodrilidae. The Lurnbricina consists of five superfaniLies. -
Different Names of Earthworm in Tamil • நாக்குப்பூச்சி
Earthworm (மண்ꯁ폁 ) Different names of earthworm in tamil நாக்埁ப்ꯂச்殿 நாங்埂ழ் ꯂநாகம் நாக்垿ளிப்ꯁ폁 நாக்垿ளிப்ꯁ폁மண்ꯁ폁 கண்டபதம் நிலவேர் ꯂ뮿வேர் மண்迁ளிப்பாம்ꯁ மண்ணில் உண்டாம் ꯁ폁 வகை வமகத்鎿ன்ힿந்鏁 உ뾿ரியல் வகைப்பா翁 鎿கண: ힿலங்垿னம் த ா埁鎿: வகை ் கைப்ꯁ폁ை்ைை் வ埁ப்ꯁ: en:Clitellata வரிகை: en:Haplotaxida 鏁கணவரிகை: Lumbricina பர்뮿毁டர், 1837 The farmer’s friend, the humble earthworm is an annelid that helps retain the fertility of the soil. This is an invertebrate animal that lives mostly in the upper layer of the soil. An earthworm is a segmented worm; a terrestrial invertebrate belonging to the phylum Annelida. They are the common inhabitants of moist soil and feed on organic matter. Earthworms are commonly called as farmer’s friend. This is because the worm casting (faecal deposit) increases the fertility and burrowing helps in proper aeration of the soil. Earthworms are commonly found in soil, eating a wide variety of organic matter includes plant matter, living protozoa, rotifers, nematodes, bacteria, fungi, and other microorganisms. Generally, within a species, the number of segments found is consistent across specimens, and individuals are born with the number of segments they will have throughout their lives. The first body segment (segment number 1) features both the earthworm's mouth and, overhanging the mouth, a fleshy lobe called the prostomium, which seals the entrance when the worm is at rest, but is also used to feel and chemically sense the worm's surroundings. Some species of earthworm can even use the prehensile prostomium to grab and drag items such as grasses and leaves into their burrow. -

Biomolecular Approach to Oligochaete Taxonomy Dr
Dr. Jaya M et. al. / International Journal of New Technologies in Science and Engineering Vol. 2, Issue 6,Dec 2015, ISSN 2349-0780 Biomolecular Approach To Oligochaete Taxonomy Dr. Jaya. M 1* ,Dr. Aja. M2 and Dr. K. Vijayakumaran Nair3 1* Assistant Professor, Department of Zoology, Sree keralavarma College, Thrissur, Email: [email protected] 2Senior Research Fellow, Department of Zoology, University of Kerala, Kariavatom, Email: [email protected] 3Associate Professor, Department of Zoology, Mar Ivanios College, Thiruvananthapuram, Email: [email protected] ABSTRACT This paper comprises the molecular approach for the identification of earthworm along with the traditional taxonomic method. The mitochondrial CO 1 gene of the Pontoscolex corethrurus (Glossoscolecidae), Travoscolides chengannures, Amynthas corticis, Perionyx sansibaricus (Megascolecidae), Progizzardus varadiamensis and Glyphidrilus annandalei (Almidae) were sequenced. The cytochrome-c oxidase I (CO1) exhibited a unique barcode to a particular species. The further exploration of mitochondrial diversity in earthworms will lead to major improvements in our understanding of the evolutionary pathways and rates of the mitochondrial genome Key words: Barcoding, cytochrome c oxidase 1 (COI), 16S ribosomal DNA INTRODUCTION DNA barcoding is a taxonomic method that uses a short genetic marker in the DNA to identify an organism. It differs from molecular phylogeny in that the main goal is to identify an unknown sample in terms of a known classification [26]. DNA sequence can be used to identify different species, in the same way as the supermarket scanner uses the black stripes of the UPC barcode to identify the items. This database will rapidly link a specimen to a Binomial Linnaean name and through that link it will provide all available information and studies on the species. -

Earthworm Ecology from DARWIN to VERMICUL TURE for 1882
Earthworm Ecology FROM DARWIN TO VERMICUL TURE FOR 1882. MAN ·I~ ·BVT ·A·woR...JV\· Frontispiece Cartoon from Punch, December 6th, 1881 Earthworm Ecology FROM DARWIN TO VERMICUL TURE Edited by J. E. Satchell Institute of Terrestrial Ecology Merlewood Research Station Grange-over-Sands Cumbria, UK LONDON NEW YORK CHAPMAN AND HALL First published 1983 by Chapman and Hall Ltd I I New Fetter Lane, London EC4P 4EE Published in the USA by Chapman and Hall 733 Third Avenue, New York NY100I7 © 1983 Chapman and Hall Ltd Softcover reprint of the hardcover 1st edition 2007 University Press, Cambridge ISBN-13: 978-94-009-5967-5 e-ISBN-13: 978-94-009-5965-1 DOl: 10.1007/978-94-009-5965-1 All rights reserved. No part ofthis book may be reprinted, or reproduced or utilized in any form or by any electronic, mechanical or other means, now known or hereafter invented, including photocopying and record ing, or in any information storage and retrieval system, without permission in writing from the Publisher. British Library Cataloguing in Publication Data Earthworm ecology. I. Opisthopora I. Satchell, J. E. 595.I' 46 QL39 I. 04 Library of Congress Cataloging in Publication Data Main entry under title: Earthworm ecology. Bibliography: p. Includes index. I. Opisthopora-Ecology. 2. Earthworm culture. I. Satchell, John E. QL39I.A6E2 7 1983 Contents Preface Xl Contributors xiii DARWIN'S CONTRIBUTION TO EARTHWORM ECOLOGY I Darwin's Formation of Vegetable Mould- its philo sophical basis M. S. Ghilarov I 2 D~rwin on earthworms - the contemporary back ground and what the critics thought O. -

External Morphological Comparison, Taxonomic Revision and Molecular Differentiation of the Four Economically Important Species of Earthworm in Thailand
INTERNATIONAL JOURNAL OF AGRICULTURE & BIOLOGY ISSN Print: 1560–8530; ISSN Online: 1814–9596 10–607/SRZ/2011/13–4–553–558 http://www.fspublishers.org Full Length Article External Morphological Comparison, Taxonomic Revision and Molecular Differentiation of the Four Economically Important Species of Earthworm in Thailand WIRIYA LOONGYAI1, PHUWADOL BANGRAK† AND SOMCHAI CHANTSAVANG Department of Animal Science, Faculty of Agriculture, Kasetsart University, Bangkok 10900 Thailand †School of Science, Walailak University, Thasala, Nakhon Si Thammarat 80160 Thailand 1Corresponding author’s e-mail: [email protected]; [email protected] ABSTRACT Four economically important species of earthworm were cultured and the external and internal characters of adult clitellate earthworms were studied. Partial sequences for ribosomal 16S rDNA and subunit one for mitochondrial cytochrome c oxidase (COI) of four earthworm species were obtained. The result of sequence analysis combined with taxonomic characters could distinguish the different species of earthworm. Morphology and nucleotide sequence of two genes for the red worm (Pheretima peguana) were distinct from Eudrilus eugeniae but were similar to the blue worm (Perionyx excavatus) and Lao worm (P. excavates) and therefore, it was classified as a new species, Perionyx sp. 1. Moreover, Eudrilus eugeniae was evidently defined as the same genus and species. Interestingly, the blue worm and Lao worm were morphologically similar to Perionyx sp. However, the molecular data of 16S rDNA could not differentiate in taxa of those two species. COI nucleotide sequence analyses showed the presence of divergent lineages between two species, suggesting the blue worm and Lao worm could be described as Perionyx sp. 2 and Perionyx sp.