Supplementary Material For
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Deciphering Molecular Mechanisms and Prioritizing Therapeutic Targets in Cardio-Oncology
Figure 1. This is a pilot view to explore the potential of EpiGraphDB to inform us about proteins that are linked to the pathophysiology of cancer and cardiovascular disease (CVD). For each cancer type (pink diamonds), we searched for cancer related proteins (light blue circles) that interact with other proteins identified as protein quantitative trait loci (pQTLs) for CVD (red diamonds for pathologies, orange triangles for risk factors). These pQTLs can be acting in cis (solid lines) or trans-acting (dotted lines). Proteins can interact either directly, a protein-protein interaction (dotted blue edges), or through the participation in the same pathway (red parallel lines). Shared pathways are represented with blue hexagons. We also queried which of these proteins are targeted by existing drugs. We found that the cancer drug cetuximab (yellow circle) inhibits EGFR. Other potential drugs are depicted in light brown hexagonal meta-nodes that are detailed below. Deciphering molecular mechanisms and prioritizing therapeutic targets in cardio-oncology Pau Erola1,2, Benjamin Elsworth1,2, Yi Liu2, Valeriia Haberland2 and Tom R Gaunt1,2,3 1 CRUK Integrative Cancer Epidemiology Programme; 2 MRC Integrative Epidemiology Unit, University of Bristol; 3 The Alan Turing Institute Cancer and cardiovascular disease (CVD) make by far the immense What is EpiGraphDB? contribution to the totality of human disease burden, and although mortality EpiGraphDB is an analytical platform and graph database that aims to is declining the number of those living with the disease shows little address the necessity of innovative and scalable approaches to harness evidence of change (Bhatnagar et al., Heart, 2016). -

Successful Chemotherapy Is Possible for Seemingly Inoperable Anaplastic Thyroid Cancer
® Clinical Thyroidology for the Public VOLUME 12 | ISSUE 12 | DECEMBER 2019 THYROID CANCER Successful chemotherapy is possible for seemingly inoperable anaplastic thyroid cancer BACKGROUND form of standard chemotherapy and 2 received another While the vast majority of thyroid cancers are slow tyrosine kinase inhibitor called pembrolizumab. Of the 6 growing and have an excellent prognosis, anaplastic patients that had surgery after this treatment, 4 patients thyroid cancer, which makes up <1% of all thyroid cancer, had the entire primary cancer removed and the other 2 is one of the most aggressive of all cancers, with a survival patients only had microscopic pieces of cancer left after averaging ~6 months after diagnosis. Surgery, radiation the surgery. After the surgery, 5 of 6 patients received and single drug chemotherapy is all ineffective in most standard chemotherapy and radiation to the surgical area. cases. The aim of this study is to study if combination Of the 6 patients, 4 patients had no evidence of cancer at chemotherapy will make previously inoperable anaplastic the last check, some over 2 years after surgery. The 2 other thyroid cancers safe to remove with surgery. patients did pass away from anaplastic cancer; however, there was no re-growth of cancer in the area where surgery THE FULL ARTICLE TITLE occurred. Wang JR et al 2019 Complete surgical resection following neoadjuvant dabrafenib plus trametinib in BRAFV600E- WHAT ARE THE IMPLICATIONS mutated anaplastic thyroid carcinoma. Thyroid 29:1036– OF THIS STUDY? 1043. PMID: 31319771. In selected patients with anaplastic thyroid cancer with the BRAF V600E mutation, treatment with dabrafenib SUMMARY OF THE STUDY and trametinib may increase the chance of having a In this study from the MD Anderson Cancer Center successful surgery of the primary tumor. -

Therapeutic Inhibition of VEGF Signaling and Associated Nephrotoxicities
REVIEW www.jasn.org Therapeutic Inhibition of VEGF Signaling and Associated Nephrotoxicities Chelsea C. Estrada,1 Alejandro Maldonado,1 and Sandeep K. Mallipattu1,2 1Division of Nephrology, Department of Medicine, Stony Brook University, Stony Brook, New York; and 2Renal Section, Northport Veterans Affairs Medical Center, Northport, New York ABSTRACT Inhibition of vascular endothelial growth factor A (VEGFA)/vascular endothelial with hypertension and proteinuria. Re- growth factor receptor 2 (VEGFR2) signaling is a common therapeutic strategy in ports describe histologic changes in the oncology, with new drugs continuously in development. In this review, we consider kidney primarily as glomerular endothe- the experimental and clinical evidence behind the diverse nephrotoxicities associ- lial injury with thrombotic microangiop- ated with the inhibition of this pathway. We also review the renal effects of VEGF athy (TMA).8 Nephrotic syndrome has inhibition’s mediation of key downstream signaling pathways, specifically MAPK/ also been observed,9 with the clinical ERK1/2, endothelial nitric oxide synthase, and mammalian target of rapamycin manifestations varying according to (mTOR). Direct VEGFA inhibition via antibody binding or VEGF trap (a soluble decoy mechanism and direct target of VEGF receptor) is associated with renal-specific thrombotic microangiopathy (TMA). Re- inhibition. ports also indicate that tyrosine kinase inhibition of the VEGF receptors is prefer- Current VEGF inhibitors can be clas- entially associated with glomerulopathies such as minimal change disease and FSGS. sifiedbytheirtargetofactioninthe Inhibition of the downstream pathway RAF/MAPK/ERK has largely been associated VEGFA-VEGFR2 pathway: drugs that with tubulointerstitial injury. Inhibition of mTOR is most commonly associated with bind to VEGFA, sequester VEGFA, in- albuminuria and podocyte injury, but has also been linked to renal-specificTMA.In hibit receptor tyrosine kinases (RTKs), all, we review the experimentally validated mechanisms by which VEGFA-VEGFR2 or inhibit downstream pathways. -

Combined BRAF and MEK Inhibition with Vemurafenib and Cobimetinib for Patients with Advanced Melanoma
Review Melanoma Combined BRAF and MEK Inhibition with Vemurafenib and Cobimetinib for Patients with Advanced Melanoma Antonio M Grimaldi, Ester Simeone, Lucia Festino, Vito Vanella and Paolo A Ascierto Melanoma, Cancer Immunotherapy and Innovative Therapy Unit, Istituto Nazionale Tumori Fondazione “G. Pascale”, Napoli, Italy cquired resistance is the most common cause of BRAF inhibitor monotherapy treatment failure, with the majority of patients experiencing disease progression with a median progression-free survival of 6-8 months. As such, there has been considerable A focus on combined therapy with dual BRAF and MEK inhibition as a means to improve outcomes compared with monotherapy. In the COMBI-d and COMBI-v trials, combined dabrafenib and trametinib was associated with significant improvements in outcomes compared with dabrafenib or vemurafenib monotherapy, in patients with BRAF-mutant metastatic melanoma. The combination of vemurafenib and cobimetinib has also been investigated. In the phase III CoBRIM study in patients with unresectable stage III-IV BRAF-mutant melanoma, treatment with vemurafenib and cobimetinib resulted in significantly longer progression-free survival and overall survival (OS) compared with vemurafenib alone. One-year OS was 74.5% in the vemurafenib and cobimetinib group and 63.8% in the vemurafenib group, while 2-year OS rates were 48.3% and 38.0%, respectively. The combination was also well tolerated, with a lower incidence of cutaneous squamous-cell carcinoma and keratoacanthoma compared with monotherapy. Dual inhibition of both MEK and BRAF appears to provide a more potent and durable anti-tumour effect than BRAF monotherapy, helping to prevent acquired resistance as well as decreasing adverse events related to BRAF inhibitor-induced activation of the MAPK-pathway. -

Original Research Paper In-Silico FDA-Approved Drug Repurposing to Find
Original Research Paper In-silico FDA-approved drug repurposing to find the possible treatment of Coronavirus Disease-19 (COVID-19) Kumar Sharp1, Dr. Shubhangi Dange2* 12nd MBBS undergraduate student, Government Medical College and Hospital, Jalgaon 2Associate Professor, Dept. of Microbiology, Government Medical College and Hospital, Jalgaon *Corresponding author: - Dr. Shubhangi Dange, Associate Professor, Department of Microbiology, Government Medical College and Hospital, Jalgaon Email: [email protected] Abstract Identification of potential drug-target interaction for approved drugs serves as the basis of repurposing drugs. Studies have shown polypharmacology as common phenomenon. In-silico approaches help in screening large compound libraries at once which could take years in a laboratory. We screened a library of 1050 FDA-approved drugs against spike glycoprotein of SARS-CoV2 in-silico. Anti-cancer drugs have shown good binding affinity which is much better than hydroxychloroquine and arbidol. We have also introduced a hypothesis named “Bump” hypothesis which and be developed further in field of computational biology. Keywords: spike glycoprotein; FDA; drug repurposing; anti-cancer; hydroxychloroquine Introduction Identification of potential drug-target interaction for approved drugs serves as the basis of repurposing drugs. Studies have shown polypharmacology as common phenomenon [1][2]. Since the three-dimensional structures of proteins of SARS-CoV2 have been mapped it opens opportunity for in-silico approaches of either novel drug discovery or drug repurposing. In the absence of an exact cure or vaccine, coronavirus disease-19 has taken a huge toll of humanity. Our study of target specific drug docking and novel hypothesis contributes in this fight. In-silico approaches help in screening large compound libraries at once which could take years in a laboratory. -

Tafinlar, INN-Dabrafenib
ANNEX I SUMMARY OF PRODUCT CHARACTERISTICS 1 1. NAME OF THE MEDICINAL PRODUCT Tafinlar 50 mg hard capsules Tafinlar 75 mg hard capsules 2. QUALITATIVE AND QUANTITATIVE COMPOSITION Tafinlar 50 mg hard capsules Each hard capsule contains dabrafenib mesilate equivalent to 50 mg of dabrafenib. Tafinlar 75 mg hard capsules Each hard capsule contains dabrafenib mesilate equivalent to 75 mg of dabrafenib. For the full list of excipients, see section 6.1. 3. PHARMACEUTICAL FORM Hard capsule (capsule). Tafinlar 50 mg hard capsules Opaque dark red capsules, approximately 18 mm long, with capsule shell imprinted with “GS TEW” and “50 mg”. Tafinlar 75 mg hard capsules Opaque dark pink capsules, approximately 19 mm long, with capsule shell imprinted with “GS LHF” and “75 mg”. 4. CLINICAL PARTICULARS 4.1 Therapeutic indications Melanoma Dabrafenib as monotherapy or in combination with trametinib is indicated for the treatment of adult patients with unresectable or metastatic melanoma with a BRAF V600 mutation (see sections 4.4 and 5.1). Adjuvant treatment of melanoma Dabrafenib in combination with trametinib is indicated for the adjuvant treatment of adult patients with Stage III melanoma with a BRAF V600 mutation, following complete resection. Non-small cell lung cancer (NSCLC) Dabrafenib in combination with trametinib is indicated for the treatment of adult patients with advanced non-small cell lung cancer with a BRAF V600 mutation. 2 4.2 Posology and method of administration Treatment with dabrafenib should be initiated and supervised by a qualified physician experienced in the use of anticancer medicinal products. Before taking dabrafenib, patients must have confirmation of tumour BRAF V600 mutation using a validated test. -

Leptomeningeal Metastases from Solid Tumors: Recent Advances in Diagnosis and Molecular Approaches
cancers Review Leptomeningeal Metastases from Solid Tumors: Recent Advances in Diagnosis and Molecular Approaches Alessia Pellerino 1,* , Priscilla K. Brastianos 2, Roberta Rudà 1,3 and Riccardo Soffietti 1 1 Department of Neuro-Oncology, University and City of Health and Science Hospital, 10126 Turin, Italy; [email protected] (R.R.); riccardo.soffi[email protected] (R.S.) 2 Massachusetts General Hospital Cancer Center, Harvard Medical School, Boston, MA 02115, USA; [email protected] 3 Department of Neurology, Castelfranco Veneto and Brain Tumor Board Treviso Hospital, 31100 Treviso, Italy * Correspondence: [email protected]; Tel.: +39-011-633-4904 Simple Summary: Leptomeningeal metastases are a devastating complication of solid tumors with poor survival, regardless of the type of treatments. The limited efficacy of targeted agents is due to the molecular divergence between leptomeningeal recurrences and primary site, as well as the presence of a heterogeneous blood-brain barrier and blood-tumor barrier that interfere with the penetration of drugs into the brain. The diagnosis of leptomeningeal metastases is achieved by neurological examination, and/or brain and spinal magnetic resonance, and/or a positive cerebrospinal fluid cytology. The presence of neoplastic cells in the cerebrospinal fluid examination is the gold-standard for the diagnosis of leptomeningeal metastases; however, novel techniques known as “liquid biopsy” aim to improve the sensitivity and specificity in detecting circulating neoplastic cells or DNA in the cerebrospinal fluid. Targeted therapies and immunotherapies have changed the natural history Citation: Pellerino, A.; Brastianos, of metastatic solid tumors, including lung, breast cancer, and melanoma. Targeting actionable P.K.; Rudà, R.; Soffietti, R. -

Oncology Drug Shortage Update
] ] [ News ] Analysis ] Commentary ] Controversy ] June 25, 2013 Vol. 35 No. 12 oncology-times.com ONCOLOGY e 5 Y ars 3 of g P in u t b a l r i s b h e i l n e g IMES C 35 TThe Independent Hem/Onc News Source Oncology Drug Shortage Update BY HEATHER LINDSEY new survey documents that cancer drug shortages have occurred A frequently, led to treatment delays, increased the risks of errors and adverse outcomes, complicated research, and increased the costs of medications� The survey, by the Hematology-Oncology Pharmacy Association, is the first to focus specifically on oncology� Page 14 • Advanced NSCLC: 60 Gy Standard • Seminoma: Opting Out of Adjuvant Therapy Safe LUNG CANCER: • Immunotherapy ‘Poised to Change Treatment Paradigm’ Leukemia Researcher Lukas Meeting Spotlights • DLBCL: CT Surveillance Adds Little Value Wartman Describes 10-Year Progress in Molecularly • CLL: Enthusiasm for Idelalisb Personal Battle Against Guided Therapy p.10 • Fitness & Cancer Prognosis pp.18, 25, 28, 33, 36, 37 Adult ALL p.20 [ALSO] SHOP TALK �� � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � �3 EDITH PEREZ: How I Treat Patients with Advanced HER2-Positive Breast Cancer � � � � � � � � � � �5 Exercise, Estrogen, & Breast Cancer Risk: New Data �� � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � �11 JOE SIMONE: Changing Views of Leaders and Leadership � � � � � � � � � � � � � � � � � � � � � � � � � � � � �16 Science Symposium for ACS’s 100th Anniversary �� � � � � � � � � � � � � � � � � � � � � � � � -

MEKINIST (Trametinib) Tablets for Oral Use Are Supplied As 0.5 Mg and 2 Mg Tablets for Oral Administration
HIGHLIGHTS OF PRESCRIBING INFORMATION Colitis and Gastrointestinal Perforation: Colitis and gastrointestinal These highlights do not include all the information needed to use perforation can occur in patients receiving MEKINIST. (5.3) MEKINIST safely and effectively. See full prescribing information for Venous Thromboembolism: Deep vein thrombosis and pulmonary MEKINIST. embolism can occur in patients receiving MEKINIST. (5.4, 2.7) MEKINIST® (trametinib) tablets, for oral use Cardiomyopathy: Assess LVEF before treatment, after one month of Initial U.S. Approval: 2013 treatment, then every 2 to 3 months thereafter. (5.5, 2.7) ------------------------------RECENT MAJOR CHANGES------------------------ Ocular Toxicities: Perform ophthalmologic evaluation for any visual Indications and Usage (1.1-1.3) 4/2018 disturbances. For Retinal Vein Occlusion (RVO), permanently Indications and Usage (1.4, 1.5) 5/2018 discontinue MEKINIST. (5.6, 2.7) Dosage and Administration Interstitial Lung disease (ILD): Withhold MEKINIST for new or (2.2, 2.3, 2.4) 4/2018 progressive unexplained pulmonary symptoms. Permanently discontinue (2.1, 2.5, 2.6, 2.7) 5/2018 MEKINIST for treatment-related ILD or pneumonitis. (5.7, 2.7) Warnings and Precautions (5) 4/2018 Serious Febrile Reactions: Can occur when MEKINIST is used with dabrafenib. (5.8, 2.7) ------------------------------INDICATIONS AND USAGE------------------------- Serious Skin Toxicity: Monitor for skin toxicities and for secondary MEKINIST is a kinase inhibitor indicated as a single agent for the treatment infections. Discontinue MEKINIST for intolerable Grade 2, or Grade 3 of patients with unresectable or metastatic melanoma with BRAF V600E or or 4 rash not improving within 3 weeks despite interruption of V600K mutations as detected by an FDA-approved test. -

Cell-Cycle and DNA-Damage Response Pathway Is Involved In
Published OnlineFirst October 13, 2017; DOI: 10.1158/1078-0432.CCR-17-1582 Biology of Human Tumors Clinical Cancer Research Cell-Cycle and DNA-Damage Response Pathway Is Involved in Leptomeningeal Metastasis of Non–Small Cell Lung Cancer Yun Fan1, Xuehua Zhu2, Yan Xu3, Xuesong Lu4, Yanjun Xu1, Mengzhao Wang3, Haiyan Xu4, Jingyan Ding2, Xin Ye2, Luo Fang5, Zhiyu Huang5, Lei Gong5, Hongyang Lu1, Weimin Mao1, and Min Hu2 Abstract Purpose: Leptomeningeal metastasis (LM) is a detrimental risk. Intriguingly, low overlapping of somatic protein-changing complication of non–small cell lung cancer (NSCLC) and asso- variants was observed between paired CSF and primary lesions, ciated with poor prognosis. However, the underlying mechanisms exhibiting tumor heterogeneity and genetic divergence. Moreover, of the metastasis process are still poorly understood. genes with CSF-recurrent genomic alterations were predominant- Experimental Design: We performed next-generation panel ly involved in cell-cycle regulation and DNA-damage response sequencing of primary tumor tissue, cerebrospinal fluid (CSF), (DDR), suggesting a role of the pathway in LM development. and matched normal controls from epidermal growth factor Conclusions: Our study has shed light on the genomic varia- receptor (EGFR) mutation-positive NSCLC patients with LM. tions of NSCLC-LM, demonstrated genetic heterogeneity and Results: The status of EGFR-activating mutations was highly divergence, uncovered involvement of cell-cycle and DDR path- concordant between primary tumor and CSF. PIK3CA aberrations way, and paved the way for potential therapeutic approaches to were high in these patients, implicating an association with LM this unmet medical need. Clin Cancer Res; 24(1); 209–16. -

First-Line Treatment Options for Patients with Stage IV Non-Small Cell Lung Cancer with Driver Alterations
First-Line Treatment Options for Patients with Stage IV Non-Small Cell Lung Cancer with Driver Alterations Patients with stage IV non-small cell lung cancer Nonsquamous cell carcinoma and squamous cell carcinoma Activating EGFR mutation other Sensitizing (L858R/exon 19 MET exon 14 skipping than exon 20 insertion mutations, EGFR exon 20 mutation ALK rearrangement ROS1 rearrangement BRAF V600E mutation RET rearrangement NTRK rearrangement mutations KRAS alterations HER2 alterations NRG1 alterations deletion) EGFR mutation T790M, L858R or Ex19Del PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options Emerging target; no Emerging target; no Emerging target; no Platinum doublet † † † Osimertinib monotherapy S Afatinib monotherapy M M Alectinib S Entrectinib M Dabrafenib/trametinib M Capmatinib M Selpercatinib M Entrectinib M conclusions available conclusions available conclusions available chemotherapy ± bevacizumab Gefitinib with doublet Standard treatment based on Standard treatment based on Standard treatment based on M M M Brigatinib S Crizotinib M M Tepotinib M Pralsetinib* W Larotrectinib M chemotherapy non-driver mutation guideline non-driver mutation guideline non-driver mutation guideline If alectinib or brigatinib are not available If entrectinib or crizotinib are not available Standard treatment based on Standard treatment based on Standard treatment based on Dacomitinib monotherapy M Osimertinib W M M M Ceritinib S Ceritinib W non-driver mutation guideline non-driver mutation guideline non-driver mutation guideline Monotherapy with afatinib M Crizotinib S Lortlatinib W Standard treatment based on Erlotinib/ramucirumab M M non-driver mutation guideline Erlotinib/bevacizumab M Monotherapy with erlotinib M Strength of Recommendation Monotherapy with gefitinib M S Strong M Moderate W Weak Monotherapy with icotinib M Notes. -
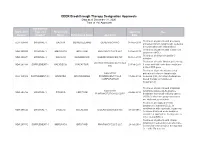
CDER Breakthrough Therapy Designation Approvals Data As of December 31, 2020 Total of 190 Approvals
CDER Breakthrough Therapy Designation Approvals Data as of December 31, 2020 Total of 190 Approvals Submission Application Type and Proprietary Approval Use Number Number Name Established Name Applicant Date Treatment of patients with previously BLA 125486 ORIGINAL-1 GAZYVA OBINUTUZUMAB GENENTECH INC 01-Nov-2013 untreated chronic lymphocytic leukemia in combination with chlorambucil Treatment of patients with mantle cell NDA 205552 ORIGINAL-1 IMBRUVICA IBRUTINIB PHARMACYCLICS LLC 13-Nov-2013 lymphoma (MCL) Treatment of chronic hepatitis C NDA 204671 ORIGINAL-1 SOVALDI SOFOSBUVIR GILEAD SCIENCES INC 06-Dec-2013 infection Treatment of cystic fibrosis patients age VERTEX PHARMACEUTICALS NDA 203188 SUPPLEMENT-4 KALYDECO IVACAFTOR 21-Feb-2014 6 years and older who have mutations INC in the CFTR gene Treatment of previously untreated NOVARTIS patients with chronic lymphocytic BLA 125326 SUPPLEMENT-60 ARZERRA OFATUMUMAB PHARMACEUTICALS 17-Apr-2014 leukemia (CLL) for whom fludarabine- CORPORATION based therapy is considered inappropriate Treatment of patients with anaplastic NOVARTIS lymphoma kinase (ALK)-positive NDA 205755 ORIGINAL-1 ZYKADIA CERITINIB 29-Apr-2014 PHARMACEUTICALS CORP metastatic non-small cell lung cancer (NSCLC) who have progressed on or are intolerant to crizotinib Treatment of relapsed chronic lymphocytic leukemia (CLL), in combination with rituximab, in patients NDA 206545 ORIGINAL-1 ZYDELIG IDELALISIB GILEAD SCIENCES INC 23-Jul-2014 for whom rituximab alone would be considered appropriate therapy due to other co-morbidities