An Endemic Rat Species Complex Is Evidence of Moderate
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Quantifying Trends of Land Change in Qinghai-Tibet Plateau During 2001–2015
remote sensing Article Quantifying Trends of Land Change in Qinghai-Tibet Plateau during 2001–2015 Chao Wang, Qiong Gao and Mei Yu * Department of Environmental Sciences, University of Puerto Rico, Rio Piedras, San Juan, PR 00936, USA; [email protected] (C.W.); [email protected] (Q.G.) * Correspondence: [email protected]; Tel.: +1-787-764-0000 Received: 1 September 2019; Accepted: 17 October 2019; Published: 20 October 2019 Abstract: The Qinghai-Tibet Plateau (QTP) is among the most sensitive ecosystems to changes in global climate and human activities, and quantifying its consequent change in land-cover land-use (LCLU) is vital for assessing the responses and feedbacks of alpine ecosystems to global climate changes. In this study, we first classified annual LCLU maps from 2001–2015 in QTP from MODIS satellite images, then analyzed the patterns of regional hotspots with significant land changes across QTP, and finally, associated these trends in land change with climate forcing and human activities. The pattern of land changes suggested that forests and closed shrublands experienced substantial expansions in the southeastern mountainous region during 2001–2015 with the expansion of massive meadow loss. Agricultural land abandonment and the conversion by conservation policies existed in QTP, and the newly-reclaimed agricultural land partially offset the loss with the resulting net change of 5.1%. Although the urban area only expanded 586 km2, mainly at the expense of agricultural − land, its rate of change was the largest (41.2%). Surface water exhibited a large expansion of 5866 km2 (10.2%) in the endorheic basins, while mountain glaciers retreated 8894 km2 ( 3.4%) mainly in the − southern and southeastern QTP. -

Study on the Coniferous Characters of Pinus Yunnanensis and Its Clustering Analysis
Journal of Polymer Science and Engineering (2017) Original Research Article Study on the Coniferous Characters of Pinus yunnanensis and Its Clustering Analysis Zongwei Zhou,Mingyu Wang,Haikun Zhao Huangshan Institute of Botany, Anhui Province, China ABSTRACT Pine is a relatively easy genus for intermediate hybridization. It has been widely believed that there should be a natural hybrid population in the distribution of Pinus massoniona Lamb. and Pinus hangshuanensis Hsia, that is, the excessive type of external form between Pinus massoniana and Pinus taiwanensis exist. This paper mainly discusses the traits and clustering analysis of coniferous lozeng in Huangshan scenic area. This study will provide a theoretical basis for the classification of long and outstanding Huangshan Song and so on. At the same time, it will provide reference for the phenomenon of gene seepage between the two species. KEYWORDS: Pinus taiwanensis Pinus massoniana coniferous seepage clustering Citation: Zhou ZW, Wang MY, ZhaoHK, et al. Study on the Coniferous Characters of Pinus yunnanensis and Its Clustering Analysis, Gene Science and Engineering (2017); 1(1): 19–27. *Correspondence to: Haikun Zhao, Huangshan Institute of Botany, Anhui Province, China, [email protected]. 1. Introduction 1.1. Research background Huangshan Song distribution in eastern China’s subtropical high mountains, more than 700m above sea level. Masson pine is widely distributed in the subtropical regions of China, at the lower reaches of the Yangtze River, vertically distributed below 700m above sea level, the upper reaches of the Yangtze River area, the vertical height of up to 1200 - 1500m or so. In the area of Huangshan Song and Pinus massoniana, an overlapping area of Huangshan Song and Pinus massoniana was formed between 700 - 1000m above sea level. -

Portfolio Investment Opportunities in China Democratic Revolution in China, Was Launched There
Morgan Stanley Smith Barney Investment Strategy The Great Wall of China In c. 220 BC, under Qin Shihuangdi (first emperor of the Qin dynasty), sections of earlier fortifications were joined together to form a united system to repel invasions from the north. Construction of the Great Wall continued for more than 16 centuries, up to the Ming dynasty (1368–1644), National Emblem of China creating the world's largest defense structure. Source: About.com, travelchinaguide.com. The design of the national emblem of the People's Republic of China shows Tiananmen under the light of five stars, and is framed with ears of grain and a cogwheel. Tiananmen is the symbol of modern China because the May 4th Movement of 1919, which marked the beginning of the new- Portfolio Investment Opportunities in China democratic revolution in China, was launched there. The meaning of the word David M. Darst, CFA Tiananmen is “Gate of Heavenly Succession.” On the emblem, the cogwheel and the ears of grain represent the working June 2011 class and the peasantry, respectively, and the five stars symbolize the solidarity of the various nationalities of China. The Han nationality makes up 92 percent of China’s total population, while the remaining eight percent are represented by over 50 nationalities, including: Mongol, Hui, Tibetan, Uygur, Miao, Yi, Zhuang, Bouyei, Korean, Manchu, Kazak, and Dai. Source: About.com, travelchinaguide.com. Please refer to important information, disclosures, and qualifications at the end of this material. Morgan Stanley Smith Barney Investment Strategy Table of Contents The Chinese Dynasties Section 1 Background Page 3 Length of Period Dynasty (or period) Extent of Period (Years) Section 2 Issues for Consideration Page 65 Xia c. -

Article-Lanzhou
13 by He Yuanqing, Yao Tandong, Cheng Guodong, and Yang Meixue Climatic records in a firn core from an Alpine temperate glacier on Mt. Yulong, southeastern part of the Tibetan Plateau Cold and Arid Regions Environmental and Engineering Research Institute, Chinese Academy of Sciences, Lanzhou 73000, China. Mt. Yulong is the southernmost glacier-covered area in Asian southwestern monsoon climate. Their total area is 11.61 km2. Eurasia, including China. There are 19 sub-tropical The glaciers resemble a group of flying dragons, giving Mt. Yulong temperate glaciers on the mountain, controlled by the (white dragon) as its name. Many explorers, tourists, poets and scientists have described southwestern monsoon climate. In the summer of 1999, Mt. Yulong from different points of view (Ward, 1924; Wissmann, a firn core, 10.10 m long, extending down to glacier ice, 1937). However, as they were unable to cross the extremely steep was recovered in the accumulation area of the largest mountain slopes and the forested area to the glacier above 4000 m a.s.l., some reported data were not correct. Most of the literature has glacier, Baishui No.1. Periodic variations of climatic focussed on the alpine landscape and the snow scenery, and there are signals above 7.8 m depth were apparent, and net accu- few accounts of the existing glaciers. Ren et al (1957) and Luo and mulation of four years was identified by the annual Yang (1963) first reported the distribution of these glaciers and of the late Pleistocene glacial landforms. To clarify the scale of glacia- oscillations of isotopic and ionic composition. -

Diversity and Geographical Pattern of Altitudinal Belts in the Hengduan Mountains in China
J. Mt. Sci. (2010) 7: 123–132 DOI: 10.1007/s11629-010-1011-9 Diversity and Geographical Pattern of Altitudinal Belts in the Hengduan Mountains in China YAO Yonghui, ZHANG Baiping*, HAN Fang, and PANG Yu Institute of Geographic Sciences and Natural Resources Research, Chinese Academy of Sciences, Beijing 100101, China. E-mail: [email protected] * Corresponding author, e-mail: [email protected] © Science Press and Institute of Mountain Hazards and Environment, CAS and Springer-Verlag Berlin Heidelberg 2010 Abstract: This paper analyses the diversity and effect needs further study. In addition, the data spatial pattern of the altitudinal belts in the quality and data accuracy at present also affect to Hengduan Mountains in China. A total of 7 types of some extent the result of quantitative modeling and base belts and 26 types of altitudinal belts are should be improved with RS/GIS in the future. identified in the study region. The main altitudinal belt lines, such as forest line, the upper limit of dark Keywords: Hengduan Mountains; Altitudinal belt coniferous forest and snow line, have similar spectra; Exposure effect; Quadratic model latitudinal and longitudinal spatial patterns, namely, arched quadratic curve model with latitudes and concave quadratic curve model along longitudinal Introduction direction. These patterns can be together called as “Hyperbolic-paraboloid model”, revealing the complexity and speciality of the environment and Globally and generally, the upper and lower ecology in the study region. This result further limits of altitudinal belts vary (increase) from high validates the hypnosis of a common quadratic model latitudes to low latitudes, from continental for spatial pattern of mountain altitudinal belts peripheries to inland areas, and from very humid proposed by the authors. -

Essd-2020-71.Pdf
Discussions https://doi.org/10.5194/essd-2020-71 Earth System Preprint. Discussion started: 7 September 2020 Science c Author(s) 2020. CC BY 4.0 License. Open Access Open Data 1 Consolidating the Randolph Glacier Inventory and the Glacier 2 Inventory of China over the Qinghai-Tibetan Plateau and th 3 Investigating Glacier Changes Since the mid-20 Century 4 Xiaowan Liu1,2,3, Zongxue Xu1,2, Hong Yang3,4, Xiuping Li5, Dingzhi Peng1,2 5 1College of Water Sciences, Beijing Normal University, Beijing, 100875, China 6 2Beijiing Key Laboratory of Urban Hydrological Cycle and Sponge City Technology, Beijing, 100875, China 7 3Eawag, Swiss Federal Institute of Aquatic Science and Technology, 8600 Dübendorf, Switzerland 8 4Department of Environmental Science, MGU University of Basel, Petersplatz 1, 4001, Switzerland 9 5Institute of Tibetan Plateau Research, Chinese Academy of Sciences, Beijing, 100101, China 10 Correspondence to: Zongxue Xu ([email protected]) 11 Abstract. Glacier retreat in the Qinghai-Tibetan Plateau (QTP), the ‘third pole of the world’, has attracted the 12 attention of researchers worldwide. Glacier inventories in the 1970s and the 2000s provide valuable information 13 to infer changes in individual glaciers. However, individual glacier volumes are either missing, incomplete or have 14 large errors in these inventories, and thus, the use of these datasets to investigate changes in glaciers in QTP in the 15 past few decades has become a challenge, particularly in the context of climate change. In this study, individual 16 glacier volume data in the Randolph Glacier Inventory version 4.0 (RGI 4.0, 1970s) and the second Glacier 17 Inventory of China (GIC-Ⅱ, 2000s) are recalculated and consolidated using a slope-dependent algorithm based on 18 elevation datasets for the QTP. -

Downloaded from Brill.Com10/07/2021 12:04:04PM Via Free Access © Koninklijke Brill NV, Leiden, 2017
Amphibia-Reptilia 38 (2017): 517-532 A new moth-preying alpine pit viper species from Qinghai-Tibetan Plateau (Viperidae, Crotalinae) Jingsong Shi1,2,∗, Gang Wang3, Xi’er Chen4, Yihao Fang5,LiDing6, Song Huang7,MianHou8,9, Jun Liu1,2, Pipeng Li9 Abstract. The Sanjiangyuan region of Qinghai-Tibetan Plateau is recognized as a biodiversity hotspot of alpine mammals but a barren area in terms of amphibians and reptiles. Here, we describe a new pit viper species, Gloydius rubromaculatus sp. n. Shi, Li and Liu, 2017 that was discovered in this region, with a brief taxonomic revision of the genus Gloydius.The new species can be distinguished from the other congeneric species by the following characteristics: cardinal crossbands on the back, indistinct canthus rostralis, glossy dorsal scales, colubrid-like oval head shape, irregular small black spots on the head scales, black eyes and high altitude distribution (3300-4770 m above sea level). The mitochondrial phylogenetic reconstruction supported the validity of the new species and furthermore reaffirms that G. intermedius changdaoensis, G. halys cognatus, G. h. caraganus and G. h. stejnegeri should be elevated as full species. Gloydius rubromaculatus sp. n. was found to be insectivorous: preying on moths (Lepidoptera, Noctuidae, Sideridis sp.) in the wild. This unusual diet may be one of the key factors to the survival of this species in such a harsh alpine environment. Keywords: Gloydius rubromaculatus sp. n., insectivorous, new species, Sanjiangyuan region. Introduction leopards (Uncia uncia), wild yaks (Bos grun- niens) and Tibetan antelopes (Pantholops hodg- The Sanjiangyuan region (the Source of Three sonii) (Shen and Tan, 2012). -

Qinghai Information
Qinghai Information Overview Qinghai is located in northwestern China. The capital and largest city, Xining, lies roughly 50 miles (80 km) from the western border and approximately 30 miles (48 km) north of the Yellow River (Huang He). It is the nation’s 4th largest province with almost 279,000 square miles (more accurately 721,000 sq km). However, the total population places 30th in the country with only 5,390,000 people. The province earns its name from the salt lake Qinghai, located in the province’s northeast less than 100 miles (161 km) west of Xining. Qinghai Lake is the largest lake in China, the word literally meaning “blue sea”. Qinghai Geography Qinghai province is located on the northeastern part of the Tibetan Plateau of western China. The Altun Mountains run along the northwestern horizontal border with Xinjiang while the Hoh Xil Mountains run horizontally over the vertical portion of that border. The Qilian Mountains run along the northeastern border with Gansu. The Kunlun Mountains follow the horizontal border between Tibet (Xizang) and Xinjiang. The Kunlun Mountains gently slope southward as the move to central Qinghai where they are extended eastward by the Bayan Har Mountains. The Dangla Mountains start in Tibet south of the Kunlun Mountains to which they run parallel. The Ningjing Mountains start in the south of Qinghai and move southward into Tibet then Yunnan. The famous Yellow River commences in this Qinghai China. A small river flows from the west into Gyaring Lake where a small outlet carries water eastward to Ngoring Lake. The Yellow River then starts on the east side of Ngoring Lake. -

Simulating the Route of the Tang-Tibet Ancient Road for One Branch of the Silk Road Across the Qinghai-Tibet Plateau
RESEARCH ARTICLE Simulating the route of the Tang-Tibet Ancient Road for one branch of the Silk Road across the Qinghai-Tibet Plateau 1 1 2 3 1 Zhuoma Lancuo , Guangliang HouID *, Changjun Xu , Yuying Liu , Yan Zhu , Wen Wang4, Yongkun Zhang4 1 Key Laboratory of Physical Geography and Environmental Process, College of Geography, Qinghai Normal University, Xining, Qinghai Province, China, 2 Key Laboratory of Geomantic Technology and Application of Qinghai Province, Provincial geomantic Center of Qinghai, Xining, Qinghai Province, China, 3 Department of a1111111111 computer technology and application, Qinghai University, Xining, Qinghai Province, China, 4 State Key a1111111111 Laboratories of Plateau Ecology and Agriculture, Qinghai University, Xining, Qinghai Province, China a1111111111 a1111111111 * [email protected] a1111111111 Abstract As the only route formed in the inner Qinghai-Tibet plateau, the Tang-Tibet Ancient Road OPEN ACCESS promoted the extension of the Overland Silk Roads to the inner Qinghai-Tibet plateau. Con- Citation: Lancuo Z, Hou G, Xu C, Liu Y, Zhu Y, sidering the Complex geographical and environmental factors of inner Qinghai-Tibet Pla- Wang W, et al. (2019) Simulating the route of the teau, we constructed a weighted trade route network based on geographical integration Tang-Tibet Ancient Road for one branch of the Silk Road across the Qinghai-Tibet Plateau. PLoS ONE factors, and then adopted the principle of minimum cost and the shortest path on the net- 14(12): e0226970. https://doi.org/10.1371/journal. work to simulate the ancient Tang-Tibet Ancient Road. We then compared the locations of pone.0226970 known key points documented in the literature, and found a significant correspondence in Editor: Wenwu Tang, University of North Carolina the Qinghai section. -
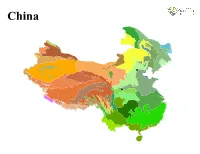
R Graphics Output
China China LEGEND Previously sampled Malaise trap site Ecoregion Alashan Plateau semi−desert North Tibetan Plateau−Kunlun Mountains alpine desert Altai alpine meadow and tundra Northeast China Plain deciduous forests Altai montane forest and forest steppe Northeast Himalayan subalpine conifer forests Altai steppe and semi−desert Northern Indochina subtropical forests Amur meadow steppe Northern Triangle subtropical forests Bohai Sea saline meadow Northwestern Himalayan alpine shrub and meadows Central China Loess Plateau mixed forests Nujiang Langcang Gorge alpine conifer and mixed forests Central Tibetan Plateau alpine steppe Ordos Plateau steppe Changbai Mountains mixed forests Pamir alpine desert and tundra Changjiang Plain evergreen forests Qaidam Basin semi−desert Da Hinggan−Dzhagdy Mountains conifer forests Qilian Mountains conifer forests Daba Mountains evergreen forests Qilian Mountains subalpine meadows Daurian forest steppe Qin Ling Mountains deciduous forests East Siberian taiga Qionglai−Minshan conifer forests Eastern Gobi desert steppe Rock and Ice Eastern Himalayan alpine shrub and meadows Sichuan Basin evergreen broadleaf forests Eastern Himalayan broadleaf forests South China−Vietnam subtropical evergreen forests Eastern Himalayan subalpine conifer forests Southeast Tibet shrublands and meadows Emin Valley steppe Southern Annamites montane rain forests Guizhou Plateau broadleaf and mixed forests Suiphun−Khanka meadows and forest meadows Hainan Island monsoon rain forests Taklimakan desert Helanshan montane conifer forests -

Records of the Transmission of the Lamp (Jingde Chuadeng
The Hokun Trust is pleased to support the fifth volume of a complete translation of this classic of Chan (Zen) Buddhism by Randolph S. Whitfield. The Records of the Transmission of the Lamp is a religious classic of the first importance for the practice and study of Zen which it is hoped will appeal both to students of Buddhism and to a wider public interested in religion as a whole. Contents Foreword by Albert Welter Preface Acknowledgements Introduction Appendix to the Introduction Abbreviations Book Eighteen Book Nineteen Book Twenty Book Twenty-one Finding List Bibliography Index Foreword The translation of the Jingde chuandeng lu (Jingde era Record of the Transmission of the Lamp) is a major accomplishment. Many have reveled in the wonders of this text. It has inspired countless numbers of East Asians, especially in China, Japan and Korea, where Chan inspired traditions – Chan, Zen, and Son – have taken root and flourished for many centuries. Indeed, the influence has been so profound and pervasive it is hard to imagine Japanese and Korean cultures without it. In the twentieth century, Western audiences also became enthralled with stories of illustrious Zen masters, many of which are rooted in the Jingde chuandeng lu. I remember meeting Alan Ginsburg, intrepid Beat poet and inveterate Buddhist aspirant, in Shanghai in 1985. He had been invited as part of a literary cultural exchange between China and the U. S., to perform a series of lectures for students at Fudan University, where I was a visiting student. Eager to meet people who he could discuss Chinese Buddhism with, I found myself ushered into his company to converse on the subject. -

Long-Term Variations in Actual Evapotranspiration Over the Tibetan Plateau
Earth Syst. Sci. Data, 13, 3513–3524, 2021 https://doi.org/10.5194/essd-13-3513-2021 © Author(s) 2021. This work is distributed under the Creative Commons Attribution 4.0 License. Long-term variations in actual evapotranspiration over the Tibetan Plateau Cunbo Han1,2,3, Yaoming Ma1,2,4,5, Binbin Wang1,2, Lei Zhong6, Weiqiang Ma1,2,4, Xuelong Chen1,2,4, and Zhongbo Su7 1Land–Atmosphere Interaction and its Climatic Effects Group, State Key Laboratory of Tibetan Plateau Earth System and Resources Environment (TPESRE), Institute of Tibetan Plateau Research, Chinese Academy of Sciences, Beijing, China 2CAS Center for Excellence in Tibetan Plateau Earth Sciences, Chinese Academy of Sciences, Beijing, China 3Institute for Meteorology and Climate Research, Karlsruhe Institute of Technology, Karlsruhe, Germany 4College of Earth and Planetary Sciences, University of Chinese Academy of Sciences, Beijing, China 5Frontier Center for Eco-environment and Climate Change in Pan-third Pole Regions, Lanzhou University, Lanzhou, China 6Laboratory for Atmospheric Observation and Climate Environment Research, School of Earth and Space Sciences, University of Science and Technology of China, Hefei, China 7Faculty of Geo-Information Science and Earth Observation, University of Twente, Enschede, the Netherlands Correspondence: Y. Ma ([email protected]) Received: 30 October 2020 – Discussion started: 11 January 2021 Revised: 7 June 2021 – Accepted: 15 June 2021 – Published: 19 July 2021 Abstract. Actual terrestrial evapotranspiration (ETa) is a key parameter controlling land–atmosphere interac- tion processes and water cycle. However, spatial distribution and temporal changes in ETa over the Tibetan Plateau (TP) remain very uncertain. Here we estimate the multiyear (2001–2018) monthly ETa and its spatial distribution on the TP by a combination of meteorological data and satellite products.