Capturing Proteins That Bind Polyunsaturated Fatty Acids: Demonstration Using Arachidonic Acid and Eicosanoids
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Wiskott-Aldrich Syndrome: the Actin Cytoskeleton and Immune Cell Function
Disease Markers 29 (2010) 157–175 157 DOI 10.3233/DMA-2010-0735 IOS Press The Wiskott-Aldrich syndrome: The actin cytoskeleton and immune cell function Michael P. Blundella, Austen Wortha,b, Gerben Boumaa and Adrian J. Thrashera,b,∗ aMolecular Immunology Unit, UCL Institute of Child Health, London, UK bDepartment of Immunology, Great Ormond Street Hospital NHS Trust, Great Ormond Street, London, UK Abstract. Wiskott-Aldrich syndrome (WAS) is a rare X-linked recessive primary immunodeficiency characterised by immune dysregulation, microthrombocytopaenia, eczema and lymphoid malignancies. Mutations in the WAS gene can lead to distinct syndrome variations which largely, although not exclusively, depend upon the mutation. Premature termination and deletions abrogate Wiskott-Aldrich syndrome protein (WASp) expression and lead to severe disease (WAS). Missense mutations usually result in reduced protein expression and the phenotypically milder X-linked thrombocytopenia (XLT) or attenuated WAS [1–3]. More recently however novel activating mutations have been described that give rise to X-linked neutropenia (XLN), a third syndrome defined by neutropenia with variable myelodysplasia [4–6]. WASP is key in transducing signals from the cell surface to the actin cytoskeleton, and a lack of WASp results in cytoskeletal defects that compromise multiple aspects of normal cellular activity including proliferation, phagocytosis, immune synapse formation, adhesion and directed migration. Keywords: Wiskott-Aldrich syndrome, actin polymerization, lymphocytes, -

Structural Basis of Thymosin-Β4/Profilin Exchange Leading to Actin Filament Polymerization
Structural basis of thymosin-β4/profilin exchange leading to actin filament polymerization Bo Xuea,1,2, Cedric Leyratb,1, Jonathan M. Grimesb,c, and Robert C. Robinsona,d aInstitute of Molecular and Cell Biology, A*STAR (Agency for Science, Technology and Research), Biopolis, Singapore 138673; bDivision of Structural Biology, Henry Wellcome Building for Genomic Medicine, Oxford, OX3 7BN, United Kingdom; cDiamond Light Source Ltd., Harwell Science and Innovation Campus, Didcot, Oxfordshire, OX11 0DE, United Kingdom; and dDepartment of Biochemistry, National University of Singapore, Singapore 117597 Edited by Thomas D. Pollard, Yale University, New Haven, CT, and approved September 22, 2014 (received for review June 30, 2014) Thymosin-β4(Tβ4) and profilin are the two major sequestering shared between Tβ4 and profilin. During phases of polymeriza- proteins that maintain the pool of monomeric actin (G-actin) within tion, the profilin–actin complex gives up its actin to the elon- cells of higher eukaryotes. Tβ4 prevents G-actin from joining a fila- gating filaments. The liberated profilin competes with Tβ4 for its ment, whereas profilin:actin only supports barbed-end elongation. bound ATP–G-actin, effectively restoring the pool of polymeri- Here, we report two Tβ4:actin structures. The first structure shows zation-competent actin. On filament dissociation, ADP-actin is that Tβ4 has two helices that bind at the barbed and pointed faces preferentially sequestered by profiling because of the afore- of G-actin, preventing the incorporation of the bound G-actin into mentioned difference in its affinity toward profilin and Tβ4. In a filament. The second structure displays a more open nucleotide the profilin:actin complex, nucleotide exchange is enhanced, and binding cleft on G-actin, which is typical of profilin:actin structures, the resulting ATP-actin is subject to competition between pro- with a concomitant disruption of the Tβ4 C-terminal helix interac- filin and Tβ4, resulting in the restoration of the pool of Tβ4- tion. -

Profilin and Formin Constitute a Pacemaker System for Robust Actin
RESEARCH ARTICLE Profilin and formin constitute a pacemaker system for robust actin filament growth Johanna Funk1, Felipe Merino2, Larisa Venkova3, Lina Heydenreich4, Jan Kierfeld4, Pablo Vargas3, Stefan Raunser2, Matthieu Piel3, Peter Bieling1* 1Department of Systemic Cell Biology, Max Planck Institute of Molecular Physiology, Dortmund, Germany; 2Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, Dortmund, Germany; 3Institut Curie UMR144 CNRS, Paris, France; 4Physics Department, TU Dortmund University, Dortmund, Germany Abstract The actin cytoskeleton drives many essential biological processes, from cell morphogenesis to motility. Assembly of functional actin networks requires control over the speed at which actin filaments grow. How this can be achieved at the high and variable levels of soluble actin subunits found in cells is unclear. Here we reconstitute assembly of mammalian, non-muscle actin filaments from physiological concentrations of profilin-actin. We discover that under these conditions, filament growth is limited by profilin dissociating from the filament end and the speed of elongation becomes insensitive to the concentration of soluble subunits. Profilin release can be directly promoted by formin actin polymerases even at saturating profilin-actin concentrations. We demonstrate that mammalian cells indeed operate at the limit to actin filament growth imposed by profilin and formins. Our results reveal how synergy between profilin and formins generates robust filament growth rates that are resilient to changes in the soluble subunit concentration. DOI: https://doi.org/10.7554/eLife.50963.001 *For correspondence: peter.bieling@mpi-dortmund. mpg.de Introduction Competing interests: The Eukaryotic cells move, change their shape and organize their interior through dynamic actin net- authors declare that no works. -

Force-Independent Interactions of Talin and Vinculin Govern Integrin-Mediated Mechanotransduction
bioRxiv preprint doi: https://doi.org/10.1101/629683; this version posted May 7, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Force-independent interactions of talin and vinculin govern integrin-mediated mechanotransduction Paul Atherton1, Franziska Lausecker1, Alexandre Carisey1, Andrew Gilmore1, David Critchley2, Igor Barsukov3, and Christoph Ballestrem1, 1Wellcome Trust Centre for Cell-Matrix Research, University of Manchester, Manchester M13 9PT, UK. 2Department of Biochemistry, University of Leicester, Lancaster Road, Leicester LE1 9HN, UK. 3Institute of Integrative Biology, University of Liverpool, BioSciences Building, Crown Street, Liverpool L69 7ZB, UK. Talin, vinculin and paxillin are core components of the dy- C-terminal D5 tail domain (Vt) that masks the talin binding namic link between integrins and actomyosin. Here we study site in the D1 domain (Cohen et al., 2005). Furthermore, a the mechanisms that mediate their activation and association FRET conformation sensor has shown that vinculin is in an using a mitochondrial-targeting assay, structure-based mutants, open conformation within FAs (Chen et al., 2005). For talin, and advanced microscopy. As expected, full-length vinculin and the primary auto-inhibitory interaction is between the F3 do- talin are auto-inhibited and do not interact with each other in main of the N-terminal FERM domain and R9, one of the this state. Contrary to previous models that propose a critical 13 -helical bundles (R1-R13) in the flexible C-terminal rod role for forces driving talin-vinculin association, our data show a that force-independent relief of auto-inhibition is sufficient to (Calderwood et al., 2013). -

Medical Cell Biology Microfilaments 1 Thomas J
MEDICAL CELL BIOLOGY MICROFILAMENTS September 24, 2003 Thomas J. Schmidt, Ph.D. Department of Physiology and Biophysics 5-610 BSB, 335-7847 Reading Assignment: Molecular Biology of the Cell (4th ed..), 2001, by B. Alberts, A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter; Chapter 16, pp. 907-925, 927-939, 943-981 Key Concepts: 1. The cytoskeleton is a complex network of protein filaments (actin filaments, intermediate filaments and microtubules) that traverses the cell cytoplasm and performs many important and diverse cellular functions. 2. Thin actin filaments, which are present in all cells, are composed of two helically interwined chains of G-actin monomers. 3. A variety of proteins including spectrin, filamin, gelsolin, thymosin, profilin, fimbrin and α-actinin regulate the dynamic state of actin filaments 4. The spectrin membrane skeleton, which is composed primarily of actin filaments located at the cytoplasmic surface of the cell membrane, is essential for maintaining cellular shape and elasticity as well as membrane stability. 5. Cell motility is mediated by actin-filaments organized into specific cellular projections referred to as lamellipodia and filopodia. Medical Cell Biology Microfilaments 1 Thomas J. Schmidt, Ph.D. Email: [email protected] September 24, 2003 Key Terms: cytoskeleton cytochalasins actin filaments (actin) phalloidins intermediate filaments (vimentin, spectrin membrane skeleton lamin) spectrin microtubules (tubulin) actin microfilaments ankyrin F-actin band 4.1 G-actin glycophorin myosin II band 3.0 myosin I hereditary spherocytosis actin microfilaments hereditary elliptocytosis treadmilling sickle cell anemia actin-binding proteins spectrin supergene family spectrin spectrin filamin α-actin fimbrin dystrophin α-actinin microvilli gelsolin terminal web thymosin lamellipodium profilin filopodia villin stress fibers contractile bundles Medical Cell Biology Microfilaments 2 Thomas J. -

New Model for Stacking Monomers in Filamentous Actin from Skeletal Muscles of Oryctolagus Cuniculus
International Journal of Molecular Sciences Article New Model for Stacking Monomers in Filamentous Actin from Skeletal Muscles of Oryctolagus cuniculus Anna V. Glyakina 1,2, Alexey K. Surin 1,3,4, Sergei Yu. Grishin 1 , Olga M. Selivanova 1, Mariya Yu. Suvorina 1, Liya G. Bobyleva 5, Ivan M. Vikhlyantsev 5 and Oxana V. Galzitskaya 1,5,* 1 Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; [email protected] (A.V.G.); [email protected] (A.K.S.); [email protected] (S.Y.G.); [email protected] (O.M.S.); [email protected] (M.Y.S.) 2 Institute of Mathematical Problems of Biology, Russian Academy of Sciences, Keldysh Institute of Applied Mathematics, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia 3 The Branch of the Institute of Bioorganic Chemistry, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia 4 State Research Center for Applied Microbiology and Biotechnology, 142279 Obolensk, Moscow Region, Russia 5 Institute of Theoretical and Experimental Biophysics, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; [email protected] (L.G.B.); [email protected] (I.M.V.) * Correspondence: [email protected]; Tel.: +7-903-675-0156 Received: 29 September 2020; Accepted: 3 November 2020; Published: 6 November 2020 Abstract: To date, some scientific evidence (limited proteolysis, mass spectrometry analysis, electron microscopy (EM)) has accumulated, which indicates that the generally accepted model of double-stranded of filamentous actin (F-actin) organization in eukaryotic cells is not the only one. This entails an ambiguous understanding of many of the key cellular processes in which F-actin is involved. -

Characterization of the Cytosolic Proteins Involved in the Amoeboid Motility of Ascaris Sperm Shawnna Marie Buttery
Florida State University Libraries Electronic Theses, Treatises and Dissertations The Graduate School 2003 Characterization of the Cytosolic Proteins Involved in the Amoeboid Motility of Ascaris Sperm Shawnna Marie Buttery Follow this and additional works at the FSU Digital Library. For more information, please contact [email protected] THE FLORIDA STATE UNIVERSITY COLLEGE OF ARTS AND SCIENCES CHARACTERIZATION OF THE CYTOSOLIC PROTEINS INVOLVED IN THE AMOEBOID MOTILITY OF ASCARIS SPERM By SHAWNNA MARIE BUTTERY A Dissertation submitted to the Department of Biological Science in partial fulfillment of the requirements for the degree of Doctor of Philosophy Degree Awarded: Fall Semester, 2003 The members of the Committee approve the dissertation of Shawnna Buttery defended on August 12, 2003. ____________________________________ Thomas M. Roberts Professor Directing Dissertation ____________________________________ Timothy A. Cross Outside Committee Member ____________________________________ Thomas C. S. Keller III Committee Member ____________________________________ Myra M. Hurt Committee Member ____________________________________ Timothy S. Moerland Committee Member Approved: __________________________________________________ Timothy S. Moerland, Chair, Department of Biological Science The Office of Graduate Studies has verified and approved the above named committee members. ii ACKNOWLEDGEMENTS This work would not have been possible without the help of many individuals throughout the years. I would like to thank Dr. Thomas Roberts for his support, guidance, and above all patience. My committee members have been a great source of useful critique and questions, for which I thank them. The members of the Roberts’ lab, both past and present, have provided a great supply of help and more importantly laughter, for that I thank: Joseph Italiano, Lawrence LeClaire, Toni Roberts, Greg Roberts, Tom Morgan, Orion Vanderlinde, Long Miao, Gail Ekman, Jean Chamoun, and Mikel Hofmann. -

Molecular Interactions of Talin and Actin: a Study on the Specific Domains of Talin Involved in the Interactions Ho-Sup Lee Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2002 Molecular interactions of talin and actin: a study on the specific domains of talin involved in the interactions Ho-Sup Lee Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Biochemistry Commons, Cell Biology Commons, and the Molecular Biology Commons Recommended Citation Lee, Ho-Sup, "Molecular interactions of talin and actin: a study on the specific domains of talin involved in the interactions " (2002). Retrospective Theses and Dissertations. 528. https://lib.dr.iastate.edu/rtd/528 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. INFORMATION TO USERS This manuscript has been reproduced from the microfilm master. UMI films the text directly from the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter face, while others may be from any type of computer printer. The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. -

Cytoskeletal Remodeling in Cancer
biology Review Cytoskeletal Remodeling in Cancer Jaya Aseervatham Department of Ophthalmology, University of Texas Health Science Center at Houston, Houston, TX 77054, USA; [email protected]; Tel.: +146-9767-0166 Received: 15 October 2020; Accepted: 4 November 2020; Published: 7 November 2020 Simple Summary: Cell migration is an essential process from embryogenesis to cell death. This is tightly regulated by numerous proteins that help in proper functioning of the cell. In diseases like cancer, this process is deregulated and helps in the dissemination of tumor cells from the primary site to secondary sites initiating the process of metastasis. For metastasis to be efficient, cytoskeletal components like actin, myosin, and intermediate filaments and their associated proteins should co-ordinate in an orderly fashion leading to the formation of many cellular protrusions-like lamellipodia and filopodia and invadopodia. Knowledge of this process is the key to control metastasis of cancer cells that leads to death in 90% of the patients. The focus of this review is giving an overall understanding of these process, concentrating on the changes in protein association and regulation and how the tumor cells use it to their advantage. Since the expression of cytoskeletal proteins can be directly related to the degree of malignancy, knowledge about these proteins will provide powerful tools to improve both cancer prognosis and treatment. Abstract: Successful metastasis depends on cell invasion, migration, host immune escape, extravasation, and angiogenesis. The process of cell invasion and migration relies on the dynamic changes taking place in the cytoskeletal components; actin, tubulin and intermediate filaments. This is possible due to the plasticity of the cytoskeleton and coordinated action of all the three, is crucial for the process of metastasis from the primary site. -

Novel Targets of Apparently Idiopathic Male Infertility
International Journal of Molecular Sciences Review Molecular Biology of Spermatogenesis: Novel Targets of Apparently Idiopathic Male Infertility Rossella Cannarella * , Rosita A. Condorelli , Laura M. Mongioì, Sandro La Vignera * and Aldo E. Calogero Department of Clinical and Experimental Medicine, University of Catania, 95123 Catania, Italy; [email protected] (R.A.C.); [email protected] (L.M.M.); [email protected] (A.E.C.) * Correspondence: [email protected] (R.C.); [email protected] (S.L.V.) Received: 8 February 2020; Accepted: 2 March 2020; Published: 3 March 2020 Abstract: Male infertility affects half of infertile couples and, currently, a relevant percentage of cases of male infertility is considered as idiopathic. Although the male contribution to human fertilization has traditionally been restricted to sperm DNA, current evidence suggest that a relevant number of sperm transcripts and proteins are involved in acrosome reactions, sperm-oocyte fusion and, once released into the oocyte, embryo growth and development. The aim of this review is to provide updated and comprehensive insight into the molecular biology of spermatogenesis, including evidence on spermatogenetic failure and underlining the role of the sperm-carried molecular factors involved in oocyte fertilization and embryo growth. This represents the first step in the identification of new possible diagnostic and, possibly, therapeutic markers in the field of apparently idiopathic male infertility. Keywords: spermatogenetic failure; embryo growth; male infertility; spermatogenesis; recurrent pregnancy loss; sperm proteome; DNA fragmentation; sperm transcriptome 1. Introduction Infertility is a widespread condition in industrialized countries, affecting up to 15% of couples of childbearing age [1]. It is defined as the inability to achieve conception after 1–2 years of unprotected sexual intercourse [2]. -

9.4 | Intermediate Filaments
354 9.4 | Intermediate Filaments The second of the three major cytoskeletal Microtubule elements to be discussed was seen in the electron microscope as solid, unbranched Intermediate filaments with a diameter of 10–12 nm. They were named in- filament termediate filaments (or IFs ). To date, intermediate filaments have only been identified in animal cells. Intermediate fila- ments are strong, flexible, ropelike fibers that provide mechani- cal strength to cells that are subjected to physical stress, Gold-labeled including neurons, muscle cells, and the epithelial cells that line anti-plectin the body’s cavities. Unlike microfilaments and microtubules, antibodies IFs are a chemically heterogeneous group of structures that, in Plectin humans, are encoded by approximately 70 different genes. The polypeptide subunits of IFs can be divided into five major classes based on the type of cell in which they are found (Table 9.2) as well as biochemical, genetic, and immunologic criteria. Figure 9.41 Cytoskeletal elements are connected to one another by We will restrict the present discussion to classes I-IV, which are protein cross-bridges. Electron micrograph of a replica of a small por- found in the construction of cytoplasmic filaments, and con- tion of the cytoskeleton of a fibroblast after selective removal of actin sider type V IFs (the lamins), which are present as part of the filaments. Individual components have been digitally colorized to assist inner lining of the nuclear envelope, in Section 12.2. visualization. Intermediate filaments (blue) are seen to be connected to IFs radiate through the cytoplasm of a wide variety of an- microtubules (red) by long wispy cross-bridges consisting of the fibrous imal cells and are often interconnected to other cytoskeletal protein plectin (green). -
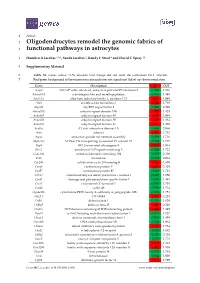
Oligodendrocytes Remodel the Genomic Fabrics of Functional Pathways in Astrocytes
1 Article 2 Oligodendrocytes remodel the genomic fabrics of 3 functional pathways in astrocytes 4 Dumitru A Iacobas 1,2,*, Sanda Iacobas 3, Randy F Stout 4 and David C Spray 2,5 5 Supplementary Material 6 Table S1. Genes whose >1.5x absolute fold-change did not meet the individual CUT criterion. 7 Red/green background of the expression ratio indicates not significant (false) up-/down-regulation. Gene Description X CUT Acap2 ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 -1.540 1.816 Adamts18 a disintegrin-like and metallopeptidase -1.514 1.594 Akr1c12 aldo-keto reductase family 1, member C12 1.866 1.994 Alx3 aristaless-like homeobox 3 1.536 1.769 Alyref2 Aly/REF export factor 2 -1.880 2.208 Ankrd33b ankyrin repeat domain 33B 1.593 1.829 Ankrd45 ankyrin repeat domain 45 1.514 1.984 Ankrd50 ankyrin repeat domain 50 1.628 1.832 Ankrd61 ankyrin repeat domain 61 1.645 1.802 Arid1a AT rich interactive domain 1A -1.668 2.066 Artn artemin 1.524 1.732 Aspm abnormal spindle microtubule assembly -1.693 1.716 Atp6v1e1 ATPase, H+ transporting, lysosomal V1 subunit E1 -1.679 1.777 Bag4 BCL2-associated athanogene 4 1.723 1.914 Birc3 baculoviral IAP repeat-containing 3 -1.588 1.722 Ccdc104 coiled-coil domain containing 104 -1.819 2.130 Ccl2 chemokine -1.699 2.034 Cdc20b cell division cycle 20 homolog B 1.512 1.605 Cenpf centromere protein F 2.041 2.128 Cep97 centrosomal protein 97 -1.641 1.723 COX1 mitochondrially encoded cytochrome c oxidase I -1.607 1.650 Cpsf7 cleavage and polyadenylation specific factor 7 -1.635 1.891 Crct1 cysteine-rich