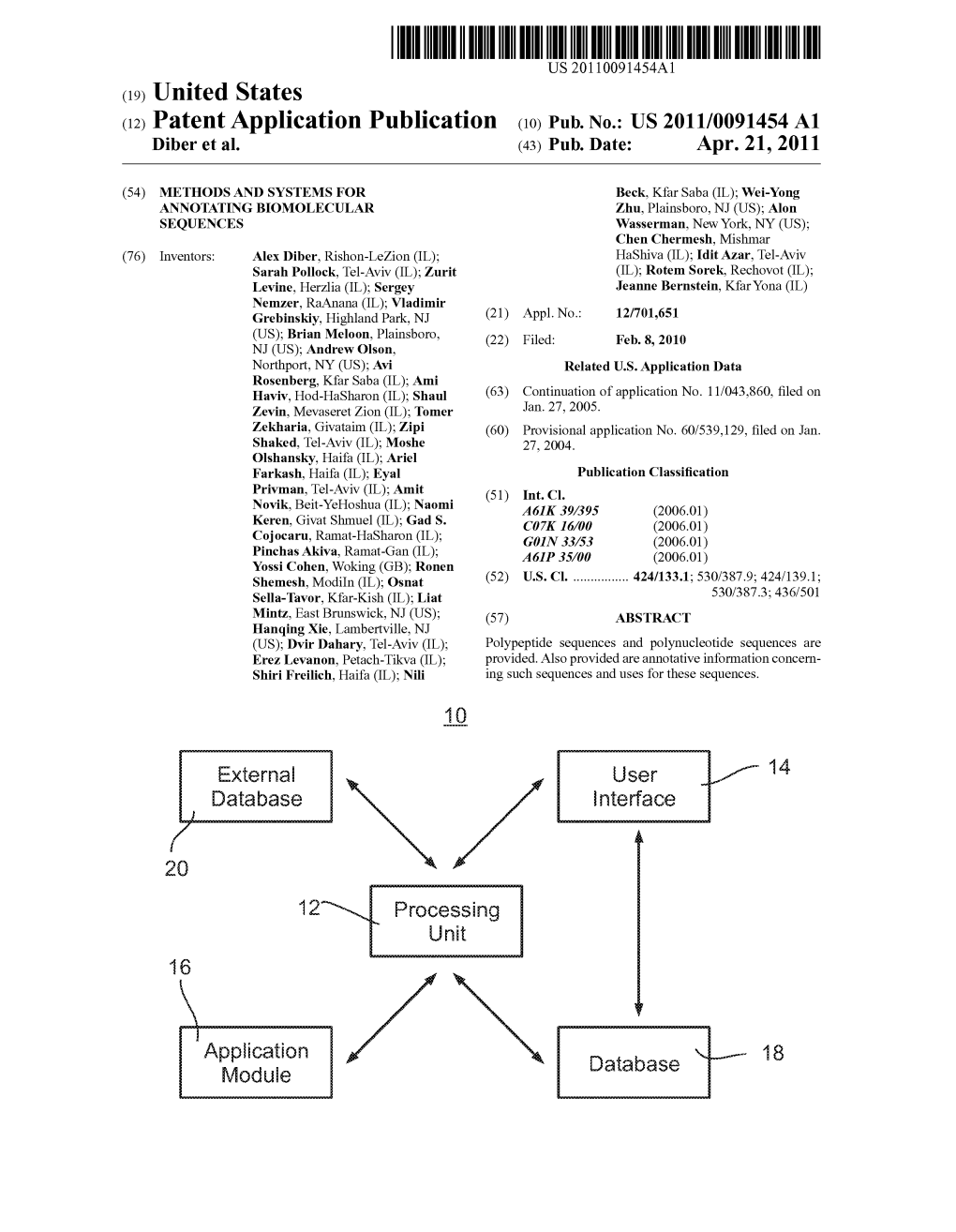
(12) Patent Application Publication (10) Pub. No.: US 2011/0091454 A1 Diber Et Al
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Insights Into Anaerobic Degradation of Benzene and Naphthalene
TECHNISCHE UNIVERSITÄT MÜNCHEN Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt Lehrstuhl für Siedlungswasserwirtschaft Insights into anaerobic degradation of benzene and naphthalene Xiyang Dong Vollständiger Abdruck der von der Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt der Technischen Universität München zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigten Dissertation. Vorsitzende(r): Prof. Dr. Wolfgang Liebl Prüfer der Dissertation: 1. Priv.-Doz. Dr. Tillmann Lueders 2. Prof. Dr. Siegfried Scherer 3. Prof. Dr. Rainer Meckenstock Die Dissertation wurde am 03.07.2017 bei der Technischen Universität München eingereicht und durch die Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt am 18.08.2017 angenommen. 天道酬勤 God rewards those who work hard Abstract Aromatic hydrocarbons, e.g. benzene and naphthalene, have toxic, mutagenic and/or carcinogenic properties. Fortunately, these compounds can be degraded in environmental systems by indigenous microorganisms. Especially under anaerobic conditions, the physiology and ecology of the microbes involved are still poorly understood. In this thesis, important knowledge gaps are addressed in this field using microbiological approaches combined with “omics tools” (metagenomics and metaproteomics). A novel “reverse stable isotope labelling” approach is also introduced to investigate biodegradation activities. Firstly, the enrichment culture BPL was studied, which can degrade benzene coupled with sulfate reduction. It is dominated by an organism of the genus Pelotomaculum. Members of this genus are usually known to be fermenters, undergoing syntrophy with anaerobic respiring microorganisms or methanogens. It remains unclear if Pelotomaculum identified here (namely, Pelotomaculum candidate BPL) could perform both benzene degradation and sulfate reduction. By using a metagenomic approach, a high-quality genome was reconstructed for it. -

Alpha-Glucan, Water Dikinase 1 Affects Starch Metabolism
www.nature.com/scientificreports OPEN Alpha-Glucan, Water Dikinase 1 Afects Starch Metabolism and Storage Root Growth in Cassava Received: 8 March 2017 Accepted: 10 August 2017 (Manihot esculenta Crantz) Published: xx xx xxxx Wenzhi Zhou1, Shutao He1, Maliwan Naconsie1, Qiuxiang Ma1, Samuel C. Zeeman2, Wilhelm Gruissem2 & Peng Zhang 1 Regulation of storage root development by source strength remains largely unknown. The cassava storage root delay (srd) T-DNA mutant postpones storage root development but manifests normal foliage growth as wild-type plants. The SRD gene was identifed as an orthologue of α-glucan, water dikinase 1 (GWD1), whose expression is regulated under conditions of light/dark cycles in leaves and is associated with storage root development. The GWD1-RNAi cassava plants showed both retarded plant and storage root growth, as a result of starch excess phenotypes with reduced photosynthetic capacity and decreased levels of soluble saccharides in their leaves. These leaves contained starch granules having greatly increased amylose content and type C semi-crystalline structures with increased short chains that suggested storage starch. In storage roots of GWD1-RNAi lines, maltose content was dramatically decreased and starches with much lower phosphorylation levels showed a drastically reduced β-amylolytic rate. These results suggested that GWD1 regulates transient starch morphogenesis and storage root growth by decreasing photo-assimilation partitioning from the source to the sink and by starch mobilization in root crops. Cassava (Manihot esculenta Crantz) is an important root crop that accumulates large amounts of starch in its storage roots, which is extensively cultivated in the tropics and subtropics1, 2. -

(12) Patent Application Publication (10) Pub. No.: US 2015/0240226A1 Mathur Et Al
US 20150240226A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2015/0240226A1 Mathur et al. (43) Pub. Date: Aug. 27, 2015 (54) NUCLEICACIDS AND PROTEINS AND CI2N 9/16 (2006.01) METHODS FOR MAKING AND USING THEMI CI2N 9/02 (2006.01) CI2N 9/78 (2006.01) (71) Applicant: BP Corporation North America Inc., CI2N 9/12 (2006.01) Naperville, IL (US) CI2N 9/24 (2006.01) CI2O 1/02 (2006.01) (72) Inventors: Eric J. Mathur, San Diego, CA (US); CI2N 9/42 (2006.01) Cathy Chang, San Marcos, CA (US) (52) U.S. Cl. CPC. CI2N 9/88 (2013.01); C12O 1/02 (2013.01); (21) Appl. No.: 14/630,006 CI2O I/04 (2013.01): CI2N 9/80 (2013.01); CI2N 9/241.1 (2013.01); C12N 9/0065 (22) Filed: Feb. 24, 2015 (2013.01); C12N 9/2437 (2013.01); C12N 9/14 Related U.S. Application Data (2013.01); C12N 9/16 (2013.01); C12N 9/0061 (2013.01); C12N 9/78 (2013.01); C12N 9/0071 (62) Division of application No. 13/400,365, filed on Feb. (2013.01); C12N 9/1241 (2013.01): CI2N 20, 2012, now Pat. No. 8,962,800, which is a division 9/2482 (2013.01); C07K 2/00 (2013.01); C12Y of application No. 1 1/817,403, filed on May 7, 2008, 305/01004 (2013.01); C12Y 1 1 1/01016 now Pat. No. 8,119,385, filed as application No. PCT/ (2013.01); C12Y302/01004 (2013.01); C12Y US2006/007642 on Mar. 3, 2006. -

Alpha-Glucan, Water Dikinase and the Control of Starch Degradation in Arabidopsis Thaliana Leaves
Alpha-Glucan, Water Dikinase and the Control of Starch Degradation in Arabidopsis thaliana Leaves Alastair William Skeffington A thesis submitted to the University of East Anglia for the degree of Doctor of Philosophy John Innes Centre Norwich September 2012 © This copy of the thesis has been supplied on condition that anyone who consults it is understood to recognise that its copyright rests with the author and that use of any information derived there-from must be in accordance with current UK Copyright Law. In addition, any quotation or extract must include full attribution. i Abstract This thesis examines the role of alpha-glucan, water dikinase (GWD) in the control of starch degradation in Arabidopsis thaliana leaves. Starch is degraded at night in Arabidopsis leaves to provide carbon for metabolism and growth. The rate of starch degradation is constant through the night and is precisely regulated such that starch reserves last until dawn. Despite the importance of this regulation for normal plant growth, we do not know which enzymes of the pathway are regulated to modulate flux. In this work I focus on the first enzyme in the starch degradation pathway, GWD. This enzyme phosphorylates the starch granule surface, thereby stimulating starch degradation by hydrolytic enzymes. It is hypothesised that GWD is important for the control of starch degradation. GWD shows large diel oscillations in transcript abundance. The enzyme is redox responsive in vitro, and is phosphorylated and SUMOylated in vivo. My work revealed that GWD protein levels are essentially invariant over the diel cycle. Studying plants in which GWD transcript was targeted by inducible RNAi and rapidly decreased to low levels revealed that GWD has half life of 2 days and a very low flux control coefficient for starch degradation. -

The Pennsylvania State University the Graduate School College Of
The Pennsylvania State University The Graduate School College of Engineering RECONSTRUCTION AND ANALYSIS OF GENOME-SCALE METABOLIC MODELS OF PHOTOSYNTHETIC ORGANISMS A Dissertation in Chemical Engineering by Rajib Saha © 2014 Rajib Saha Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy August 2014 The dissertation of Rajib Saha was reviewed and approved* by the following: Costas D. Maranas Donald B. Broughton Professor of Chemical Engineering Dissertation Advisor Chair of Committee Reka Albert Professor of Physics and Biology Howard M. Salis Assistant Professor in Chemical Engineering and Agricultural and Biological Engineering Andrew Zydney Walter L. Robb Chair and Professor of Chemical Engineering Head of the department of Chemical Engineering *Signatures are on file in the Graduate School iii ABSTRACT The scope and breadth of genome-scale metabolic reconstructions has continued to expand over the last decade. However, only a limited number of efforts exist on photosynthetic metabolism reconstruction. Cyanobacteria are an important group of photoautotrophic organisms that can synthesize valuable bio-products by harnessing solar energy. They are endowed with high photosynthetic efficiencies and diverse metabolic capabilities that confer the ability to convert solar energy into a variety of biofuels and their precursors. However, less well studied are the similarities and differences in metabolism of different species of cyanobacteria as they pertain to their suitability as microbial production chassis. Here we assemble, update and compare genome-scale models (iCyt773 and iSyn731) for two phylogenetically related cyanobacterial species, namely Cyanothece sp. ATCC 51142 and Synechocystis sp. PCC 6803. Comparisons of model predictions against gene essentiality data reveal a specificity of 0.94 (94/100) and a sensitivity of 1 (19/19) for the Synechocystis iSyn731 model.