Identification of a Novel Parvovirus in Domestic Cats
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

“Please You Cannot Ignore Any Longer the Fact That These Animals Have
NAME AFFILIATION Diana Reiss, Ph.D. New York Aquarium and Columbia University “Please you Lori Marino, Ph.D. Emory University Prof. PatholoGy, School of Medicine, Sam RidGway, D.V.M., Ph.D. cannot ignore University of California Director of the New York Aquarium Paul J. Boyle, Ph.D. any longer the and The Ocean Project fact that these Randall Wells, Ph.D. Chicago ZooloGical Society animals have Hal Whitehead, Ph.D. Dalhousie University, Canada Louis M. Herman, Ph.D. University of Hawaii very large Paul NachtiGall, Ph.D. University of Hawaii brains, highly Richard Connor. Ph.D. University of Massachusetts, Dartmouth Adjunct Professor, Scripps Institution of William F. Perrin, Ph.D. developed OceanoGraphy Executive Vice Provost, Florida International societies, social DouGlas Wartzok, Ph.D. University relationships and Robin W. Baird, Ph.D. Cascadia Research Collective sophisticated Christopher W. Clark, Ph.D. Cornell University cognitive Adam Pack, Ph.D. University of Hawaii William E. Evans, Ph.D. University of Notre Dame abilities.” James Estes, Ph.D. Adjunct Professor, UC Santa Cruz – Richard Connor, Ph.D., University of Massachusetts Laureline Meynier, Ph.D. Massey University, New Zealand David Bain University of WashinGton Center for Marine Biodiversity and Sarah Mesnick, Ph.D. Conservation Janet Mann, Ph.D. GeorGetown University Leszek Karczmarski, Ph.D. Texas A&M University Corrie Allen, Ph.D. Lone Wolf Bioscience Giovanni Di Guardo. DVM, University of Teramo, Italy Dipl. ECVP Dr. L.P. Verstraten University of Amsterdam Cornelis Hazevoet Ph.D. Lisbon National Museum of Natural History Giuseppe Notarbartolo di Tethys Research Institute, Italy Sciara Ph.D. Dr. Victoria Turner Appin Scientific Limited Heidi Lyn, Ph.D. -

Table of Contents
Table of Contents Welcome Message from the General Chair.......................................................... 2 M&N 2019 Organizers .......................................................................................... 3 Sponsors .............................................................................................................. 6 Patrons ................................................................................................................. 7 Exhibitors ............................................................................................................ 10 Monday, July 8 ....................................................................................................12 Tuesday, July 9 ................................................... ................................................19 Wednesday, July 10 ........................................................................................... 26 1 Welcome Message from the General Co-Chairs Dear colleagues and friends, On behalf of the entire Conference Committee, we are pleased to welcome you to the 5th IEEE International Symposium on Measurements and Networking (M&N 2019), which is held in Catania and hosted in Museo Diocesano in the heart of the city. The Symposium is mainly promoted by the IEEE IMS TC-37 Measurements and Networking, the IEEE IM Italy Chapter and by the IEEE Italy Section Systems Council Chapter. IEEE M&N is a privileged forum for the discussion of current and emerging trends on measurements, communications, computer science, -

Molecular Analysis of Carnivore Protoparvovirus Detected in White Blood Cells of Naturally Infected Cats
Balboni et al. BMC Veterinary Research (2018) 14:41 DOI 10.1186/s12917-018-1356-9 RESEARCHARTICLE Open Access Molecular analysis of carnivore Protoparvovirus detected in white blood cells of naturally infected cats Andrea Balboni1, Francesca Bassi1, Stefano De Arcangeli1, Rosanna Zobba2, Carla Dedola2, Alberto Alberti2 and Mara Battilani1* Abstract Background: Cats are susceptible to feline panleukopenia virus (FPV) and canine parvovirus (CPV) variants 2a, 2b and 2c. Detection of FPV and CPV variants in apparently healthy cats and their persistence in white blood cells (WBC) and other tissues when neutralising antibodies are simultaneously present, suggest that parvovirus may persist long-term in the tissues of cats post-infection without causing clinical signs. The aim of this study was to screen a population of 54 cats from Sardinia (Italy) for the presence of both FPV and CPV DNA within buffy coat samples using polymerase chain reaction (PCR). The DNA viral load, genetic diversity, phylogeny and antibody titres against parvoviruses were investigated in the positive cats. Results: Carnivore protoparvovirus 1 DNA was detected in nine cats (16.7%). Viral DNA was reassembled to FPV in four cats and to CPV (CPV-2b and 2c) in four cats; one subject showed an unusually high genetic complexity with mixed infection involving FPV and CPV-2c. Antibodies against parvovirus were detected in all subjects which tested positive to DNA parvoviruses. Conclusions: The identification of FPV and CPV DNA in the WBC of asymptomatic cats, despite the presence of specific antibodies against parvoviruses, and the high genetic heterogeneity detected in one sample, confirmed the relevant epidemiological role of cats in parvovirus infection. -
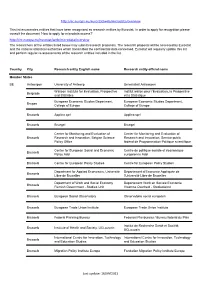
Eurostat: Recognized Research Entity
http://ec.europa.eu/eurostat/web/microdata/overview This list enumerates entities that have been recognised as research entities by Eurostat. In order to apply for recognition please consult the document 'How to apply for microdata access?' http://ec.europa.eu/eurostat/web/microdata/overview The researchers of the entities listed below may submit research proposals. The research proposal will be assessed by Eurostat and the national statistical authorities which transmitted the confidential data concerned. Eurostat will regularly update this list and perform regular re-assessments of the research entities included in the list. Country City Research entity English name Research entity official name Member States BE Antwerpen University of Antwerp Universiteit Antwerpen Walloon Institute for Evaluation, Prospective Institut wallon pour l'Evaluation, la Prospective Belgrade and Statistics et la Statistique European Economic Studies Department, European Economic Studies Department, Bruges College of Europe College of Europe Brussels Applica sprl Applica sprl Brussels Bruegel Bruegel Center for Monitoring and Evaluation of Center for Monitoring and Evaluation of Brussels Research and Innovation, Belgian Science Research and Innovation, Service public Policy Office fédéral de Programmation Politique scientifique Centre for European Social and Economic Centre de politique sociale et économique Brussels Policy Asbl européenne Asbl Brussels Centre for European Policy Studies Centre for European Policy Studies Department for Applied Economics, -

UNIVERSITY of TERAMO (Italy) * * * * *
“International Co-operation Against Trans-national Financial Organized Crime” Master – University of Teramo UNIVERSITY OF TERAMO (Italy) * * * * * CALL FOR APPLICATIONS MASTER IN “I NTERNATIONAL CO-OPERATION AGAINST TRANS -NATIONAL FINANCIAL ORGANIZED CRIME ” Academic Year 2009-2010 DURATION 2. environmental crimes, such as unlawful 1 year divided in three parts: building, pollution, trafficking in waste, Part 1 : starting from Spring 2010 at the University of radioactive slag, animals’ and plants’ Teramo, consisting in 200 hours of didactic activity. endangered species The didactic activity will be held with the following 3. corruption and bribery schedule: Thursday afternoon, Friday morning and 4. brand and copyright counterfeiting afternoon and Saturday morning; this calendar might 5. pharmaceutical counterfeiting and food undergo variations. adulteration 6. money laundering and financial crime Part 2 : starting after the end of part 1 an internship 7. terrorism and terrorism financing with international or national relevant institutions, or 8. terrorism and biotechnologies a research programme with scientific institutions. 9. cyber crime Admission to the internship will be granted to those and the substantive action undertaken by the who have attended at least 80% of the didactic activity following national and international organisations: hours and passed the intermediary written - DNA ( Direzione Nazionale Antimafia ) examinations. - EUROJUST ( European Union’s Judicial The internship period is not compulsory for Cooperation Unit ) – Permanent Mission of Italy candidates. - EUROPOL Part 3 : Candidates that have successfully concluded - INTERPOL their internship period must elaborate and defend a - OCSE ( Organisation for Economic Co-operation final thesis (not less than 100 pages). and Development ) - Permanent Mission of Italy Candidates not attending an internship must elaborate - OLAF ( European Anti-Fraud Office ) and discuss a final thesis (not less than 300 pages). -

Porcine Parvovirus VP1/VP2 on a Time Series Epitope Mapping: Exploring the Effects of High Hydrostatic Pressure on the Immune Recognition of Antigens
bioRxiv preprint doi: https://doi.org/10.1101/330589; this version posted May 25, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Porcine Parvovirus VP1/VP2 on a Time Series Epitope Mapping: exploring the effects of high hydrostatic pressure on the immune recognition of antigens. Ancelmo Rabelo de Souzaa, Marriam Yamina, Danielle Gavac, Janice Reis Ciacci Zanellac, Maria Sílvia Viccari Gattia, Carlos Francisco Sampaio Bonafea, Daniel Ferreira de Lima Netoa,b* aDepartamento de Bioquímica e Biologia Tecidual e bDepartamento de Genética, Evolução e Bioagentes, Instituto de Biologia, Universidade Estadual de Campinas (UNICAMP), Rua Monteiro Lobato, 255, Cidade Universitária Zeferino Vaz, 13083- 862, Campinas, SP, Brazil. cEmbrapa Suínos e Aves, Laboratório de Virologia de Suínos, 89715-899, Concórdia, SC, Brazil. *Corresponding author: Tel.: +55 19 3521-6229; E-mail: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/330589; this version posted May 25, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. ABSTRACT Porcine parvovirus (PPV) is a DNA virus that causes reproductive failure in gilts and sows, resulting in embryonic and fetal losses worldwide. Epitope mapping of PPV is important for developing new vaccines. In this study, we used spot synthesis analysis for epitope mapping of the capsid proteins of PPV (NADL-2 strain) and correlated the findings with predictive data from immunoinformatics. The virus was exposed to three conditions prior to inoculation in pigs: native (untreated), high hydrostatic pressure (350 MPa for 1 h) at room temperature and high hydrostatic pressure (350 MPa for 1 h) at -18 °C, compared with a commercial vaccine produced using inactivated PPV. -

67Th Economic Policy Panel
67th Economic Policy Panel Hosted by the Swiss National Bank Venue: Great Guild Hall, 2nd Floor, Zunfthaus zur Zimmerleuten Limmatquai 40, 8001 Zurich, Switzerland 12-13 April 2018 Programme Each session is 70 minutes in duration Author: 25 mins | Discussant: 15 mins (each) | Panel discussion: 15 mins * indicates presenting author Thursday 12 April 14:00 Registration and coffee on arrival 14:30 – 14:45 Opening Remarks Mr Thomas Moser, Alternate Member of the Governing Board, Swiss National Bank 14:45 – 15:55 Can Education Compensate the Effect of Population Aging on Macroeconomic Performance? Evidence from Panel Data Rainer Kotschy (LMU Munich) * Uwe Sunde (LMU Munich) Discussants: Francesco Drago (University of Messina) Pietro Biroli (University of Zurich) 15:55 – 17:05 Monetary Policy and Bank Profitability in a Low Interest Rate Environment * Carlo Altavilla (European Central Bank) Miguel Boucinha (European Central Bank) José-Luis Peydró (ICREA-UPF, CREI & BGSE) Discussants: Ralph De Haas (EBRD) Vasso Ioannidou (University of Lancaster) 17:05 – 17:20 coffee break 17:20 – 18:30 The Walking Dead?: Zombie Firms and Productivity Performance in OECD Countries Müge Adalet McGowan (OECD) Dan Andrews (OECD) * Valentine Millot (OECD) Discussants: Martin Brown (University of St.Gallen) Elena Carletti (Bocconi University) 20:00 Conference dinner at The Ballroom of the Savoy Hotel Baur en Ville Address: Poststrasse 12, 8001 Zurich Welcome address by Ms Andréa M. Maechler, Member of the Governing Board, Swiss National Bank Friday 13 April 08:30 – 09:40 Where Do People Get Their News? * Patrick Kennedy (Columbia University) Andrea Prat (Columbia University) Discussants: Roberto Galbiati (Sciences Po) David Hémous (University of Zurich) 09:40 – 10:50 Populism and Civil Society * Tito Boeri (Bocconi University) Prachi Mishra (IMF) Chris Papageorgiou (IMF) Antonio Spilimbergo (IMF) Discussants: Christina Gathmann (Heidelberg University) 10:50 – 11:10 Coffee break 11:10 – 12:20 The Gains from Economic Integration David Comerford (University of Strathclyde) * José V. -

Youth Forum 11-12 July, Trieste, ITALY
The following is the list of signatories of the present DECLARATION : 1 Agricultural University of Tirana Albania 2 University of Elbasan Albania 3 Graz University of Technology Austria 4 University of Banja Luka Bosnia and Herzegovina 5 University ‘D zˇemal Bijedi c´’ Mostar Bosnia and Herzegovina 6 University of Mostar Bosnia and Herzegovina 7 University of Split Croatia 8 University of Zadar Croatia 9 Juraj Dobrila University of Pula Croatia 10 Technological Educational Institute of Epirus Greece 11 University of Ioannina Greece 12 Ionian University Greece 13 University of Patras Greece 14 University of Bologna Italy 15 University of Camerino Italy 16 Technical University of Marche Italy TRIESTE 17 University of Trieste Italy 18 University of Udine Italy 19 University of Urbino Italy 20 University of Campania Italy 21 University of Genua Italy 22 University of Foggia Italy DECLARATION 23 University of Insubria Italy 24 University of Modena and Reggio Emilia Italy 25 University of Naples Italy 26 University of Piemonte Orientale Italy 27 University of Teramo Italy 28 University of Palermo Italy 29 University of Milano-Bicocca Italy 30 University of Tuscia Italy 31 University of Venice Ca’Foscari Italy 32 International School for Advanced Studies Italy 33 L’Orientale University of Naples Italy 34 IMT School for Advanced Studies Lucca Italy 35 University of Montenegro Montenegro 36 University of Oradea Romania 37 University Politehnica of Bucharest Romania 38 West University of Timisoara Romania 39 University of Arts in Belgrade Serbia -

Protoparvovirus Knocking at the Nuclear Door
viruses Review Protoparvovirus Knocking at the Nuclear Door Elina Mäntylä 1 ID , Michael Kann 2,3,4 and Maija Vihinen-Ranta 1,* 1 Department of Biological and Environmental Science and Nanoscience Center, University of Jyvaskyla, FI-40500 Jyvaskyla, Finland; elina.h.mantyla@jyu.fi 2 Laboratoire de Microbiologie Fondamentale et Pathogénicité, University of Bordeaux, UMR 5234, F-33076 Bordeaux, France; [email protected] 3 Centre national de la recherche scientifique (CNRS), Microbiologie Fondamentale et Pathogénicité, UMR 5234, F-33076 Bordeaux, France 4 Centre Hospitalier Universitaire de Bordeaux, Service de Virologie, F-33076 Bordeaux, France * Correspondence: maija.vihinen-ranta@jyu.fi; Tel.: +358-400-248-118 Received: 5 September 2017; Accepted: 29 September 2017; Published: 2 October 2017 Abstract: Protoparvoviruses target the nucleus due to their dependence on the cellular reproduction machinery during the replication and expression of their single-stranded DNA genome. In recent years, our understanding of the multistep process of the capsid nuclear import has improved, and led to the discovery of unique viral nuclear entry strategies. Preceded by endosomal transport, endosomal escape and microtubule-mediated movement to the vicinity of the nuclear envelope, the protoparvoviruses interact with the nuclear pore complexes. The capsids are transported actively across the nuclear pore complexes using nuclear import receptors. The nuclear import is sometimes accompanied by structural changes in the nuclear envelope, and is completed by intranuclear disassembly of capsids and chromatinization of the viral genome. This review discusses the nuclear import strategies of protoparvoviruses and describes its dynamics comprising active and passive movement, and directed and diffusive motion of capsids in the molecularly crowded environment of the cell. -

March 14-15, 2002 Federal Reserve Bank of Atlanta
TENTH ANNUAL SYMPOSIUM OF THE SOCIETY FOR NONLINEAR DYNAMICS AND ECONOMETRICS March 14-15, 2002 Federal Reserve Bank of Atlanta THURSDAY, MARCH 14 8:00 A.M. - 8:45 A.M. REGISTRATION AND CONTINENTAL BREAKFAST 8:45 A.M. - 9:00 A.M. WELCOMING REMARKS 9:00 A.M. - 10:30 A.M. FINANCE I Chair: Gerald Dwyer “Order Time, Multiple Shocks, and Short Selling in Security Price Adjustment” Malay K. Dey (Morgan State University) “The Interaction of Speculation and Diversification” Roberto Dieci (University of Parma) “Asset Pricing with a Continuum of Belief Types” Cees Diks (University of Amsterdam) Roy van der Weide (University of Amsterdam) “Convergence and Biases of Monte Carlo Estimates of American Option Prices Using a Parametric Exercise Rule” Diego Garcia (Dartmouth College) 10:30 A.M. - 11:00 A.M. BREAK 11:00 A.M. - 12:30 P.M. TIME SERIES I Chair: James Ramsey “Do Long Swings in the Business Cycle Lead to Strong Persistence in Output?” Mark Jensen (Brigham Young University) Ming Liu (University of Missouri) “Identification of Coefficients in a Quadratic Moving Average Process Using the Generalized Method of Moments” Richard A. Ashley (Virginia Tech) Douglas M. Patterson (Virginia Tech) “An ARMA Representation of Unobserved Component Models under Generalized Random Walk Specifications: New Algorithms and Examples Marcos Bujosa (Universidad Complutense de Madrid) Antonio Garcia-Ferrer (Universidad Autonoma de Madrid) Peter Young (Lancaster University) “Perturbation Solution of Nonlinear Rational Expectations Models” Peter A. Zadrozny (Bureau of Labor Statistics) Baoline Chen (Rutgers University-Camden) 12:30 P.M. - 2:00 P.M. LUNCH 2:00 P.M. -

Biomass and Waste Gasification Country Report ITALY IEA Bioenergy Task 33
Biomass and Waste Gasification Country Report ITALY IEA Bioenergy Task 33 2018 Front cover information panel IEA Bioenergy: Task 33: 2019: 11 Biomass and Waste Gasification Country Report ITALY IEA Bioenergy Task 33 2018 Donatella Barisano, ENEA (Agenzia nazionale per le nuove tecnologie, l'energia e lo sviluppo economico sostenibile) Copyright © 2015 IEA Bioenergy. All rights Reserved ISBN, if applicable, here Published by IEA Bioenergy IEA Bioenergy, also known as the Technology Collaboration Programme (TCP) for a Programme of Research, Development and Demonstration on Bioenergy, functions within a Framework created by the International Energy Agency (IEA). Views, findings and publications of IEA Bioenergy do not necessarily represent the views or policies of the IEA Secretariat or of its individual Member countries. TABLE OF CONTENTS LIST OF FIGURES ....................................................................................................................... 5 LIST OF TABLES ......................................................................................................................... 7 INTRODUCTION ......................................................................................................................... 8 RD&D PROGRAMMES TO PROMOTE THE RENEWABLE ENERGY SECTOR. .................................... 8 THE ITALIAN SUPPORT TO ELECTRICITY PRODUCTION FROM BIOMASS .................................. 8 DIFFUSION OF GASIFICATION TECHNOLOGY ON THE NATIONAL TERRITORY: THE PLANTS CONNECTED TO THE GRID. ..................................................................................................... -

MARILENA BAZZANO Educational Activity 2020-2021
CURRICULUM VITAE – MARILENA BAZZANO Educational Activity 2020-2021 - Lecturer in Veterinary Internal Medicine Practical Activities (1 CFU), School of Veterinary Medicine, University of Camerino (UNICAM). 2020-2021 – Lecturer in Veterinary Obstetric Techniques (5 CFU), Post-Graduate School of Animal Health, Breeding and Production, University of Camerino (UNICAM) 2020-2021 – Lecturer in Control of Diseases in Livestock Production Systems (5 CFU), Animal Production Science and Valorization of Animal Derived Food, University of Camerino (UNICAM). 2019-2020 – Lecturer in Control of Diseases in Livestock Production Systems (5 CFU), Animal Production Science and Valorization of Animal Derived Food, University of Camerino (UNICAM). 2019-2020 - Lecturer in Veterinary Internal Medicine Practical Activities (3 CFU), School of Veterinary Medicine, University of Camerino (UNICAM). 2019-2020 – Lecturer in Veterinary Obstetric Techniques (10 CFU), Post-Graduate School of Animal Health, Breeding and Production, University of Camerino (UNICAM) 2018-2019 – Lecturer in Veterinary Sport Medicine (3 CFU), School of Veterinary Medicine, University of Camerino (UNICAM). 2018-2019 – Lecturer in Veterinary Internal Medicine Practical Activities (3 CFU), School of Veterinary Medicine, University of Camerino (UNICAM). 2018-2019 – Lecturer in Veterinary Obstetric Techniques (10 CFU), Post-Graduate School of Animal Health, Breeding and Production, University of Camerino (UNICAM) Work Experience and Education 2019-2020 – Tutor UNICAM for Post-Graduate grant of Dr. Marta Florence David in cooperation With the School of Veterinary Medicine of the University of Liege. 2017-2019 – Co-Supervisor of PhD course in Life and Health Sciences: One Health. XXXI Cycle. School of Advanced Studies, university of Camerino. Thesis “Study of a neW formulation of functional food for dogs With chronic renal and cardiovascular diseases”, PhD student Dr.