By USDA APHIS BRS Document Control Officer at 2:18 Pm, Dec 11
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Disability Classification System
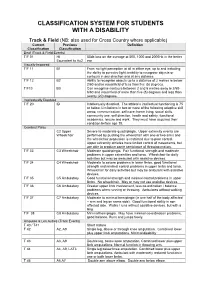
CLASSIFICATION SYSTEM FOR STUDENTS WITH A DISABILITY Track & Field (NB: also used for Cross Country where applicable) Current Previous Definition Classification Classification Deaf (Track & Field Events) T/F 01 HI 55db loss on the average at 500, 1000 and 2000Hz in the better Equivalent to Au2 ear Visually Impaired T/F 11 B1 From no light perception at all in either eye, up to and including the ability to perceive light; inability to recognise objects or contours in any direction and at any distance. T/F 12 B2 Ability to recognise objects up to a distance of 2 metres ie below 2/60 and/or visual field of less than five (5) degrees. T/F13 B3 Can recognise contours between 2 and 6 metres away ie 2/60- 6/60 and visual field of more than five (5) degrees and less than twenty (20) degrees. Intellectually Disabled T/F 20 ID Intellectually disabled. The athlete’s intellectual functioning is 75 or below. Limitations in two or more of the following adaptive skill areas; communication, self-care; home living, social skills, community use, self direction, health and safety, functional academics, leisure and work. They must have acquired their condition before age 18. Cerebral Palsy C2 Upper Severe to moderate quadriplegia. Upper extremity events are Wheelchair performed by pushing the wheelchair with one or two arms and the wheelchair propulsion is restricted due to poor control. Upper extremity athletes have limited control of movements, but are able to produce some semblance of throwing motion. T/F 33 C3 Wheelchair Moderate quadriplegia. Fair functional strength and moderate problems in upper extremities and torso. -

High-Throughput Sequencing of Six Bamboo Chloroplast Genomes: Phylogenetic Implications for Temperate Woody Bamboos (Poaceae: Bambusoideae)
High-Throughput Sequencing of Six Bamboo Chloroplast Genomes: Phylogenetic Implications for Temperate Woody Bamboos (Poaceae: Bambusoideae) Yun-Jie Zhang1,2,3., Peng-Fei Ma1,2,3., De-Zhu Li1,2* 1 Key Laboratory of Biodiversity and Biogeography, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan, People’s Republic of China, 2 Plant Germplasm and Genomics Center, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan, People’s Republic of China, 3 Graduate University of Chinese Academy of Sciences, Beijing, People’s Republic of China Abstract Background: Bambusoideae is the only subfamily that contains woody members in the grass family, Poaceae. In phylogenetic analyses, Bambusoideae, Pooideae and Ehrhartoideae formed the BEP clade, yet the internal relationships of this clade are controversial. The distinctive life history (infrequent flowering and predominance of asexual reproduction) of woody bamboos makes them an interesting but taxonomically difficult group. Phylogenetic analyses based on large DNA fragments could only provide a moderate resolution of woody bamboo relationships, although a robust phylogenetic tree is needed to elucidate their evolutionary history. Phylogenomics is an alternative choice for resolving difficult phylogenies. Methodology/Principal Findings: Here we present the complete nucleotide sequences of six woody bamboo chloroplast (cp) genomes using Illumina sequencing. These genomes are similar to those of other grasses and rather conservative in evolution. We constructed a phylogeny of Poaceae from 24 complete cp genomes including 21 grass species. Within the BEP clade, we found strong support for a sister relationship between Bambusoideae and Pooideae. In a substantial improvement over prior studies, all six nodes within Bambusoideae were supported with $0.95 posterior probability from Bayesian inference and 5/6 nodes resolved with 100% bootstrap support in maximum parsimony and maximum likelihood analyses. -

Glufosinate-Tolerant Cotton: Tolerance and Weed
GLUFOSINATE-TOLERANT COTTON: TOLERANCE AND WEED MANAGEMENT by LESLI KRISTEN BLAIR, B.S. A THESIS IN CROP SCIENCE Submitted to the Graduate Faculty of Texas Tech University in Partial Fulfillment of the Requirements for the Degree of MASTER OF SCIENCE Approved Accepted Deamof the Graduate School December, 1991 ^^•ft:;^ ACKNOWLEDGEMENTS 1i would like to extend my heartfelt gratitude to the members that served on 7 '^ '^* my advisory committee for their time, effort, and assistance. I would like to thank Dr. Dotray for his guidance and direction in the preparation of this thesis; Dr. Keeling for his instruction and support in monitoring these experiments; Dr. Gannaway for his knowledge and assistance in the biotechnology and breeding associated with this project; and Dr. Thompson for her friendship and support in making career decisions. Without all of their guidance, I could never have achieved the goals that we set. I wish to thank all of my fellow graduate students in Weed Science for their help in completing this project. I would especially like to thank Alan Helm for his endless help collecting all of the first year data, Ginger Light for her help in clarifying concepts, and LeAnna Lyon for her help with the last year's project. Their support, friendship, and help on this project have been immeasurable. I would like to express my sincerest thanks to AgrEvo and TxCot for their funding of this research. My thanks also go to the people at the USDA-ARS in Lubbock for all of their help before and after I became involved with this research. -

WO 2013/037955 Al 21 March 2013 (21.03.2013) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2013/037955 Al 21 March 2013 (21.03.2013) P O P C T (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every A01N 25/00 (2006.01) A OIN 43/653 (2006.01) kind of national protection available): AE, AG, AL, AM, A0 41/06 (2006.01) A01N 37/50 (2006.01) AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, (21) Number: International Application DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, PCT/EP2012/068096 HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, (22) International Filing Date: KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, 14 September 2012 (14.09.2012) ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, (25) Filing Language: English RW, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, (26) Publication Language: English TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: 1118 1702.9 16 September 201 1 (16.09.201 1) EP (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (71) Applicant (for all designated States except US): BAYER GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, INTELLECTUAL PROPERTY GMBH [DE/DE]; Al- UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, fred-Nobel-Str. -

Instruction Manual
EN7528563-00 07 - 2017 LGN power DYNACIAT Instruction manual EN CONTENTS PAGE 1 INTRODUCTION 2 15 INSTALLATION ANTIFREEZE PROTECTION 13 2 SHIPMENT OF THE UNIT 3 16 ELECTRICAL CONNECTIONS 13 3 RECEIPT OF GOODS 3 16.1 Power connection 13 3.1 Checking the equipment 3 16.2 Connection to the air-cooled condenser. 14 3.2 Identifying the equipment 3 16.3 Customer connection for remote 4 SAFETY INSTRUCTIONS 4 control functions. 15 5 MACHINE COMPLIANCE 4 17 CONTROL AND SAFETY DEVICES 16 17.1 Electronic control and display module 16 6 WARRANTY 4 17.2 Main functions 16 7 UNIT LOCATION 4 17.3 Safety device management 16 8 HANDLING AND POSITIONING 5 17.4 Phase controller kit 17 17.5 Location of the safety sensors and devices 18 9 LOCATION 6 17.6 Adjusting the control and safety devices 19 9.1 Location of the unit 6 18 COMMISSIONING 19 10 OPERATING LIMITS 7 18.1 Commissioning 19 10.1 Operating range 7 18.2 Essential points that must be checked 20 10.2 Limits 7 19 TECHNICAL CHARACTERISTICS 21 10.3 Evaporator limits 7 10.4 Minimum/maximum water flow rates 7 20 ELECTRICAL SPECIFICATIONS 22 11 LOCATION OF THE MAIN COMPONENTS 8 21. SERVICING AND MAINTENANCE 23 12 MAIN COMPONENTS OF THE 21.1 Operating readings 23 REFRIGERATING CIRCUIT 8 21.2 Unit maintenance and servicing 23 13 HYDRAULIC CONNECTIONS 9 22 ECODESIGN 25 13.1 Diameters of the hydraulic connections 9 23 PERMANENT SHUTDOWN 25 13.2 Connections 10 24 TROUBLESHOOTING OPERATING 13.3 FLANGE/VICTAULIC adapter kit (OPTION) 10 PROBLEMS 26 14 REFRIGERANT CONNECTIONS 11 25 SYSTEM SCHEMATICS 27 14.1 General information 11 25.1 ooling installation with drycooler 27 14.2 Accessories 11 14.3 Routing and sizing 11 14.4 Installation 12 ORIGINAL TEXT: FRENCH VERSION EN - 1 1 INTRODUCTION DYNACIATpower LGN series water chillers are designed to meet the air conditioning requirements of residential and office buildings as well as the requirements of manufacturing processes. -

INDEX to PESTICIDE TYPES and FAMILIES and PART 180 TOLERANCE INFORMATION of PESTICIDE CHEMICALS in FOOD and FEED COMMODITIES
US Environmental Protection Agency Office of Pesticide Programs INDEX to PESTICIDE TYPES and FAMILIES and PART 180 TOLERANCE INFORMATION of PESTICIDE CHEMICALS in FOOD and FEED COMMODITIES Note: Pesticide tolerance information is updated in the Code of Federal Regulations on a weekly basis. EPA plans to update these indexes biannually. These indexes are current as of the date indicated in the pdf file. For the latest information on pesticide tolerances, please check the electronic Code of Federal Regulations (eCFR) at http://www.access.gpo.gov/nara/cfr/waisidx_07/40cfrv23_07.html 1 40 CFR Type Family Common name CAS Number PC code 180.163 Acaricide bridged diphenyl Dicofol (1,1-Bis(chlorophenyl)-2,2,2-trichloroethanol) 115-32-2 10501 180.198 Acaricide phosphonate Trichlorfon 52-68-6 57901 180.259 Acaricide sulfite ester Propargite 2312-35-8 97601 180.446 Acaricide tetrazine Clofentezine 74115-24-5 125501 180.448 Acaricide thiazolidine Hexythiazox 78587-05-0 128849 180.517 Acaricide phenylpyrazole Fipronil 120068-37-3 129121 180.566 Acaricide pyrazole Fenpyroximate 134098-61-6 129131 180.572 Acaricide carbazate Bifenazate 149877-41-8 586 180.593 Acaricide unclassified Etoxazole 153233-91-1 107091 180.599 Acaricide unclassified Acequinocyl 57960-19-7 6329 180.341 Acaricide, fungicide dinitrophenol Dinocap (2, 4-Dinitro-6-octylphenyl crotonate and 2,6-dinitro-4- 39300-45-3 36001 octylphenyl crotonate} 180.111 Acaricide, insecticide organophosphorus Malathion 121-75-5 57701 180.182 Acaricide, insecticide cyclodiene Endosulfan 115-29-7 79401 -

TPS23861 IEEE 802.3At Quad Port Power-Over-Ethernet PSE
Product Order Technical Tools & Support & Folder Now Documents Software Community TPS23861 SLUSBX9I –MARCH 2014–REVISED JULY 2019 TPS23861 IEEE 802.3at Quad Port Power-over-Ethernet PSE Controller 1 Features 3 Description The TPS23861 is an easy-to-use, flexible, 1• IEEE 802.3at Quad Port PSE controller IEEE802.3at PSE solution. As shipped, it – Auto Detect, classification automatically manages four 802.3at ports without the – Auto Turn-On and disconnect need for any external control. – Efficient 255-mΩ sense resistor The TPS23861 automatically detects Powered • Pin-Out enables Two-Layer PCB Devices (PDs) that have a valid signature, determines • Kelvin Current Sensing power requirements according to classification and applies power. Two-event classification is supported • 4-Point detection for type-2 PDs. The TPS23861 supports DC • Automatic mode – as shipped disconnection and the external FET architecture – No External terminal setting required allows designers to balance size, efficiency and – No Initial I2C communication required solution cost requirements. • Semi-Automatic mode – set by I2C command The unique pin-out enables 2-layer PCB designs via logical grouping and clear upper and lower – Continuous Identification and Classification differentiation of I2C and power pins. This delivers – Meets IEEE 400-ms TPON specification best-in-class thermal performance, Kelvin accuracy – Fast-Port shutdown input and low-build cost. – Operates best when used in conjunction with In addition to automatic operation, the TPS23861 system reference code supports Semi-Auto Mode via I2C control for precision http://www.ti.com/product/TPS23861/toolssoftw monitoring and intelligent power management. are Compliance with the 400-ms TPON specification is ensured whether in semi-automatic or automatic • Optional I2C control and monitoring mode. -

2019 NFCA Texas High School Leadoff Classic Main Bracket Results
2019 NFCA Texas High School Leadoff Classic Main Bracket Results Bryan College Station BHS CSHS 1 Bryan College Station 17 Thu 3pm Thu 3pm Cedar Creek BHS CSHS EP Eastlake SA Brandeis 49 Brandeis College Station 57 Cy Woods 1 BHS Thu 5pm Thu 5pm CSHS 2 18 Thu 1pm Brandeis Robinson Thu 1pm EP Montwood BHS CSHS Robinson 11 Huntsville 3 89 Klein Splendora 93 Splendora 9 BHS Fri 2pm Fri 2pm CSHS 3 Fredericksburg Splendora 19 Thu 9am Thu 9am Fredericksburg 6 VET1 VET2 Belton 6 Grapevine 50 58 Richmond Foster 11 BHS Thu 5pm Klein Splendora Thu 5pm CSHS 4 20 Thu 11am Klein Richmond Foster Thu 11am Klein 11 BHS CSHS Rockwall 2 SA Southwest 9 97 SA Southwest Cedar Ridge 99 Cy Ranch 7 VET1 Fri 4pm Fri 4pm CP3 5 Southwest Cy Ranch 21 Thu 11am Thu 1pm Temple 5 VET1 CP3 Plano East 5 Clear Springs 2 51 Southwest Alvin 59 Alvin VET1 Thu 3pm Thu 5pm CP3 6 22 Thu 9am Vandegrift Alvin Thu 3pm Vandegrift 5 BHS CSHS Lufkin San Marcos 5 90 94 Flower Mound 0 VET2 Fri 12pm Southwest Cedar Ridge Fri 12pm CP4 7 San Marcos Cedar Ridge 23 Thu 9am Thu 1pm Magnolia West 3 VET2 CHAMPIONSHIP CP4 RR Cedar Ridge 12 McKinney Boyd 3 52 Game 192 60 SA Johnson VET2 Thu 3pm San Marcos BHS Cedar Ridge Thu 5 pm CP4 8 4:00 PM 24 Thu 11am M. Boyd BHS 5 10 BHS Johnson Thu 3pm Manvel 0 129 Southwest vs. Cedar Ridge 130 Kingwood Park Bellaire 3 Sat 10am Sat 8am Deer Park VET4 BRAC-BB 9 Cedar Park Deer Park 25 Thu 11am Thu 1pm Cedar Park 5 VET4 BRAC-BB Leander Clements 13 53 Cedar Park MacArthur 61 SA MacArthur VET4 Thu 3pm Thu 5pm BRAC-BB 10 26 Thu 9am Clements MacArthur Thu 3pm Waco University 0 BHS CSHS Tomball Memorial Cy Fair 4 91 Friendswood Woodlands 95 Woodlands 2 VET5 Fri 8am Fri 8am BRAC-YS 11 San Benito Woodlands 27 Thu 11am Thu 1pm San Benito 10 VET5 BRAC-YS SA Holmes 1 Friendswood 10 54 62 Santa Fe VET5 Thu 3pm Friendswood Woodlands Thu 5pm BRAC-YS 12 28 Thu 9am Friendswood Santa Fe Thu 3pm Henderson 0 VET1 VET2 Lake Travis Ridge Point 16 98 100 B. -

Phylogeny and Subfamilial Classification of the Grasses (Poaceae) Author(S): Grass Phylogeny Working Group, Nigel P
Phylogeny and Subfamilial Classification of the Grasses (Poaceae) Author(s): Grass Phylogeny Working Group, Nigel P. Barker, Lynn G. Clark, Jerrold I. Davis, Melvin R. Duvall, Gerald F. Guala, Catherine Hsiao, Elizabeth A. Kellogg, H. Peter Linder Source: Annals of the Missouri Botanical Garden, Vol. 88, No. 3 (Summer, 2001), pp. 373-457 Published by: Missouri Botanical Garden Press Stable URL: http://www.jstor.org/stable/3298585 Accessed: 06/10/2008 11:05 Your use of the JSTOR archive indicates your acceptance of JSTOR's Terms and Conditions of Use, available at http://www.jstor.org/page/info/about/policies/terms.jsp. JSTOR's Terms and Conditions of Use provides, in part, that unless you have obtained prior permission, you may not download an entire issue of a journal or multiple copies of articles, and you may use content in the JSTOR archive only for your personal, non-commercial use. Please contact the publisher regarding any further use of this work. Publisher contact information may be obtained at http://www.jstor.org/action/showPublisher?publisherCode=mobot. Each copy of any part of a JSTOR transmission must contain the same copyright notice that appears on the screen or printed page of such transmission. JSTOR is a not-for-profit organization founded in 1995 to build trusted digital archives for scholarship. We work with the scholarly community to preserve their work and the materials they rely upon, and to build a common research platform that promotes the discovery and use of these resources. For more information about JSTOR, please contact [email protected]. -

WO 2013/020985 Al 14 February 2013 (14.02.2013) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2013/020985 Al 14 February 2013 (14.02.2013) P O P C T (51) International Patent Classification: (74) Agent: BAYER INTELLECTUAL PROPERTY A 47/06 (2006.01) A01P 7/02 (2006.01) GMBH; Alfred-Nobel-Str. 10, 40789 Monheim (DE). A01N 57/20 (2006.01) A01P 7/04 (2006.01) (81) Designated States (unless otherwise indicated, for every A01P 5/00 (2006.01) kind of national protection available): AE, AG, AL, AM, (21) International Application Number: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, PCT/EP2012/065469 BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (22) International Filing Date: HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, 7 August 2012 (07.08.2012) KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, (25) Filing Language: English ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, (26) Publication Language: English SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, (30) Priority Data: TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, 61/521,827 10 August 201 1 (10.08.201 1) US ZW. (71) Applicant (for all designated States except US): BAYER (84) Designated States (unless otherwise indicated, for every INTELLECTUAL PROPERTY GMBH [DE/DE]; Al- kind of regional protection available): ARIPO (BW, GH, fred-Nobel-Str. -

Publication 938 15:26 - 30-MAR-2011
Userid: SD_5PQGB DTD tipx Leadpct: 0% Pt. size: 8 J Draft J Ok to Print PAGER/SGML Fileid: ...uments_1\Pub_938\4th Quarter_2010\P938_4Qtr_RPG_Final_Ron.xml (Init. & date) Page 1 of 100 of Publication 938 15:26 - 30-MAR-2011 The type and rule above prints on all proofs including departmental reproduction proofs. MUST be removed before printing. Publication 938 Introduction (Rev. March 2011) Section references are to the Internal Revenue Department Cat. No. 10647L Code unless otherwise noted. of the This publication contains directories relating Treasury to real estate mortgage investment conduits Internal (REMICs) and collateralized debt obligations Revenue Real Estate (CDOs). The directory for each calendar quarter Service is based on information submitted to the IRS during that quarter. This publication is only avail- Mortgage able on the Internet. For each quarter, there is a directory of new REMICs and CDOs, and a section containing Investment amended listings. You can use the directory to find the representative of the REMIC or the is- suer of the CDO from whom you can request tax Conduits information. The amended listing section shows changes to previously listed REMICs and CDOs. The update for each calendar quarter will (REMICs) be added to this publication approximately six weeks after the end of the quarter. Reporting Other information. Publication 550, Invest- ment Income and Expenses, discusses the tax treatment that applies to holders of these invest- ment products. For other information about Information REMICs, see sections 860A through 860G of the Internal Revenue Code (IRC) and any regu- lations issued under those sections. (And Other Publication 938 is only available electroni- cally. -

Octopine Synthase Mrna Isolated from Sunflower Crown Gall Callus Is
Proc. Nati Acad. Sci. USA Vol. 79, pp. 86-90, January 1982 Biochemistry Octopine synthase mRNA isolated from sunflower crown gall callus is homologous to the Ti plasmid of Agrobacterium tumefaciens (recombinant plasmid/hybridization/mRNA size/in vitro translation/immunoprecipitation) NORIMOTO MURAI AND JOHN D. KEMP Department of Plant Pathology, University ofWisconsin, and Plant Disease Research Unit, ARS, USDA, Madison, Wisconsin 53706 Communicated by Folke Skoog, September 14, 1981 ABSTRACT We have shown that the structural gene for oc- families have been identified. The enzymes responsible for the topine synthase (a crown gall-specific enzyme) is located in a cen- synthesis of the first two have been purified and characterized tral portion ofthe T-DNA that came from the Ti plasmid ofAgro- (17, 18). bacterium tumefaciens and is expressed after it has been trans- The particular opine family present in a crown gall cell cor- ferred to the plant cells. Polyadenylylated RNA was prepared relates with a particular Ti plasmid rather than with the host from polysomes isolated from an octopine-producing crown gall plant (19, 20). Analysis ofdeletion mutants of an octopine-type callus and purified by selective hybridization to one offive recom- Ti plasmid has shown that a gene controlling octopine produc- binant plasmids. Each such plasmid contained a different frag- tion is located on a 14.6-kbp fragment ofthe T-DNA generated ment ofT-DNA ofpTi-15955 (octopine-type Ti plasmid). Purified by Sin 1 (21) (Fig. 1). Detailed examination ofthe physical or- mRNA was translated in vitro in rabbit reticulocyte lysates, and ganization of T-DNA in octopine-type tumor lines has shown the translation products were immunoprecipitated with antibody that octopine production is correlated with the presence ofthe against octopine synthase.