Differences in Fine-Scale Genetic Structure and Dispersal In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Oaks (Quercus Spp.): a Brief History
Publication WSFNR-20-25A April 2020 Oaks (Quercus spp.): A Brief History Dr. Kim D. Coder, Professor of Tree Biology & Health Care / University Hill Fellow University of Georgia Warnell School of Forestry & Natural Resources Quercus (oak) is the largest tree genus in temperate and sub-tropical areas of the Northern Hemisphere with an extensive distribution. (Denk et.al. 2010) Oaks are the most dominant trees of North America both in species number and biomass. (Hipp et.al. 2018) The three North America oak groups (white, red / black, and golden-cup) represent roughly 60% (~255) of the ~435 species within the Quercus genus worldwide. (Hipp et.al. 2018; McVay et.al. 2017a) Oak group development over time helped determine current species, and can suggest relationships which foster hybridization. The red / black and white oaks developed during a warm phase in global climate at high latitudes in what today is the boreal forest zone. From this northern location, both oak groups spread together southward across the continent splitting into a large eastern United States pathway, and much smaller western and far western paths. Both species groups spread into the eastern United States, then southward, and continued into Mexico and Central America as far as Columbia. (Hipp et.al. 2018) Today, Mexico is considered the world center of oak diversity. (Hipp et.al. 2018) Figure 1 shows genus, sub-genus and sections of Quercus (oak). History of Oak Species Groups Oaks developed under much different climates and environments than today. By examining how oaks developed and diversified into small, closely related groups, the native set of Georgia oak species can be better appreciated and understood in how they are related, share gene sets, or hybridize. -

Victoria-Park-Tree-Walk-2-Web.Pdf
Opening times Victoria Park was London’s first The park is open every day except Christmas K public ‘park for the people’. K Day 7.00 am to dusk. Please be aware that R L Designed in 1841 by James A closing times fluctuate with the seasons. The P A specific closing time for the day of your visit is Pennethorne, it covers 88 hectares A I W listed on the park notice boards located at and contains over 4,500 trees. R E O each entrance. Trees are the largest living things on E T C Toilets are opened daily, from 10.00 am until R the planet and Victoria Park has a I V T one hour before the park is closed. variety of interesting specimens, Getting to the park many of which are as old as the park itself. Whatever the season, as you Bus: 277 Grove Road, D6 Grove Road, stroll around take time to enjoy 8 Old Ford Road their splendour, whether it’s the Tube: Mile End, Bow Road, Bethnal Green regimental design of the formal DLR: Bow Church tree-lined avenues, the exotic trees Rail: Hackney Wick (BR North London Line) from around the world or, indeed West Walk the evidence of the destruction caused by the great storm of 1987 that reminds us of the awesome power of nature. The West Walk is one of three Victoria Park tree walks devised by Tower Hamlets Council. We hope you enjoy your visit, if you have any comments or questions about trees please contact the Arboricultural department on 020 7364 7104. -

Quercus Drymeja Unger and Q. Mediterranea Unger
Review of Palaeobotany and Palynology 241 (2017) 98–128 Contents lists available at ScienceDirect Review of Palaeobotany and Palynology journal homepage: www.elsevier.com/locate/revpalbo Taxonomy and palaeoecology of two widespread western Eurasian Neogene sclerophyllous oak species: Quercus drymeja Unger and Q. mediterranea Unger Thomas Denk a,⁎, Dimitrios Velitzelos b,TuncayH.Günerc, Johannes M. Bouchal a,d, Friðgeir Grímsson d,GuidoW.Grimmd,e a Swedish Museum of Natural History, Department of Palaeobiology, Box 50007, 10405 Stockholm, Sweden b National and Kapodistrian University of Athens, Faculty of Geology and Geoenvironment, Department of Historical Geology and Paleontology, Panepistimiopolis, Athens 15784, Greece c Istanbul University, Faculty of Forestry, Department of Forest Botany, 34473 Bahceköy, Istanbul, Turkey d University of Vienna, Department of Palaeontology, 1090 Vienna, Austria e Unaffiliated, 45100 Orléans, France article info abstract Article history: Sclerophyllous oaks (genus Quercus) play important roles in Neogene ecosystems of south-western Eurasia. Received 31 May 2016 Modern analogues (‘nearest living relatives’) for these oaks have been sought among five of six infrageneric lin- Accepted 30 January 2017 eages of Quercus, distributed across the entire Northern Hemisphere. A revision of leaf fossils from lower Miocene Available online 10 February 2017 to Pliocene deposits suggests that morphotypes of the Quercus drymeja complex are very similar to a number of extant Himalayan, East Asian, and Southeast Asian species of Quercus Group Ilex and may indicate subtropical, Keywords: Quercus Group Ilex relatively humid conditions. Quercus mediterranea comprises leaf morphotypes that are encountered in modern Plant fossil Mediterranean species of Quercus Group Ilex, but also in Himalayan and East Asian members of this group indi- Modern analogue cating fully humid or summer-wet conditions. -

Mycological Notes 11: Boletus Edulis in Canterbury Jerry Cooper, July 2012
Mycological Notes 11: Boletus edulis in Canterbury Jerry Cooper, July 2012 Probably most mycologically minded New Zealanders know about Boletus edulis in Christchurch. Wang et al (1995) reported its occurrence with a number of host trees across the City and a single collection from Lake Pukaki with Birch. A well-known stronghold is with Quercus robur (Oak) in Hagley Park where it occurs with a range of other fungi. Wang et al speculate that B. edulis arrived with early European settlers into Christchurch and spread, perhaps with nursery plants in some cases. Research by Stringer et al (2002 & pers.comm), indicates that the old oak trees in Hagley Park arrived as living plants in half-barrels on-board ship. Thus the soil+fungus+roots+tree were shipped together. Even the possible source of the nursery near Bagshot in the UK has been traced. Similar shipments were apparently made to Nelson and planted in parks there. Recently there are reports of the fungus from various plantations in North Canterbury. In New Zealand the bolete is relatively immune to attack by fungous gnats, unlike its home range where most specimens rapidly become a maggot nursery. In recent years I have noticed more damage, so either the associated fungus gnat has arrived, or local species have developed a taste for it. The fungus has also been introduced/arrived with Pinus plantations in South Africa, Zimbabwe, and Chile. Wang et al also recognised that the name ‘Boletus edulis’ has been used in a broad sense by some, with morphological/host differences recognised as separate species by others. -

Geographic Distribution of 24 Major Tree Species
TECHNICAL REPORT Maximize the production of goods and services by Mediterranean forest ecosystems in a context of global changes January 2015 Geographic distribution of 24 major tree species in the Mediterranean and their genetic resources This report is the result of work conducted by the Secretariat of the FAO Silva Mediterranea Committee and Plan Bleu as part of the project ”Maximize the production of goods and services of Mediterranean forest ecosystems in the context of global changes”, funded by the French Global Environment Facility (FFEM) for the period 2012-2016. LEGAL NOTICE The designations emplyoyed and the presentation of material in this information product do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organi- zation of the United Nations (FAO) or Plan Bleu pour l’Environnememnt et le Développement en Méditerranée (Plan Bleu) concerning the legal or development status of any country, territory, city or area or of its authorities, whther or not these have been patented, does not imply that these have been endorsed or recommended by FAO or Plan Bleu in preference to others of a similar nature that are not mentioned. The views expressed in this information product are those of the author(s) and do not necessarily reflect the views or policies of FAO or Plan Bleu. COPYRIGHT This publication may be reproduced in whole or in part of any form fro educational or non-profit purposes without special permission from the copyright holder, provided akcnowledgment of the source is made. FAO would appreciat receiving a copy of any publication that uses his publication as a source. -
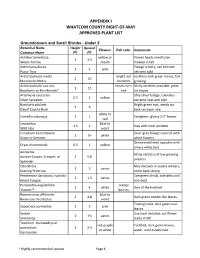
Whatcom County's Approved Plant List
APPENDIX I WHATCOM COUNTY RIGHT-OF-WAY APPROVED PLANT LIST Groundcovers and Small Shrubs - Under 2' Botanical Name Height Spread Flowers Fall color Comments Common Name (ft) (ft) Achillea tomentosa yellow or Flower heads need to be 1 3-5 Wooly Yarrow cream mowed in fall Antennaria dioica Foliage is furry, can tolerate 1 2 pink Pussy Toes extreme cold Arctostaphylos media bright red Leathery dark green leaves, fast 2 10 Manzanita Media branches growing Arctostaphylos uva ursi leaves turn Many varieties available, great 1 15 Bearberry or Kinnikinnick* red for slopes Artemesia caucasica Silky silver foliage, tolerates 0.5 2 yellow Silver Spreader extreme heat and cold Baccharis pilularis Bright green mat, needs cut 2 6 Dwarf Coyote Bush back once per year white to Camellia sasanqua 1 1 Evergreen, glossy 2-3" leaves red Ceanothus blue to 1.5 2 Stay with local varieties Wild Lilac violet Cerastium tomentosum Silver gray foliage covered with 1 5+ white Snow-in-Summer white flowers Ornamental seed capsules with Dryas drummondii 0.5 1 yellow silvery white tails Juniperus Many varieties of low growing Juniper-Carpet, Creeper, or 2 5-8 junipers Spreader Oenothera May die back in severe winters, 1 2 varies Evening Primrose come back strong Penstemon davidsonii, rupicola Evergreen shrub, tolerates cold 2 1.5 varies Beard Tongue and heat Pyracantha augustifolia orange 1 4 white One of the hardiest 'Gnome'* berries Rosmariunus officinalis blue to 2 4-8 Dark green needle-like leaves Rosemary 'Prostratus' violet Trailing habit, dark green oval Saponaria ocymoides 1 3 pink leaves Sedum Use local varieties, cut flower 2 1-5 varies Stonecrop stalks in fall Teucrium chamaedrys or red-purple Toothed, dark green leaves, postratium 1 2-3 or white water until established Germander * Highly-recommended species Page 1 Thymus white to 2 1-2 Many varieties Thyme purple Shrubs Botanical Name Height Spread Flowers Fall color Comments Common Name (ft) (ft) Arctostaphylos bright red Shiny dark green leaves turn 2-15 20 varies Manzanita berries maroon in fall. -

The Scented Isle
Corsica - The Scented Isle Naturetrek Tour Report 30 April - 7 May 2017 Corsican Fire Salamander Group at Col de Sevi Long-lipped Tongue-orchid Woodlark Report & images compiled by Andrew Bray Naturetrek Mingledown Barn Wolf's Lane Chawton Alton Hampshire GU34 3HJ UK T: +44 (0)1962 733051 E: [email protected] W: www.naturetrek.co.uk Tour Report Corsica - The Scented Isle Tour participants: Andrew Bray & Richard Lansdown (leaders) with 11 Naturetrek clients Day 1 Sunday 30th April After a flight from London Gatwick we arrived at Bastia airport, where Tongue Orchids were outside the entrance. Arranging the vehicles took a little longer than expected, but once sorted we were on our way, heading to Ponte Leccia. Here we had a coffee stop and Andrew bought some cheese, meat and bread. We then headed across the top of the island to l’Ile-Rousse, where we stopped at the farmer’s market to buy fruit and salad. We then pushed on to our lunch stop near Galeria. Here we saw a variety of birds, wall lizards and some endemic plants: Corsican Storksbill (Erodium corsicum) and Sea Lavender (Limonium corsicum). We then drove a few hundred yards to see if there was a way down to some wetland, but unfortunately there was not. We did hear Cetti’s Warbler and saw a pair of Long-tailed Tits. Our next stop was on the coastal road at one of the U bends at the head of one of the many valleys we had to negotiate. Here were even more endemic plants, though we stopped for the Wild Vine (Vitis riparia) and saw the Illyrian Sea Lily (Pancratium Illyricum). -

Resilience of Sweet Chestnut and Truffle Holm-Oak Rural Forests in Languedoc-Roussillon, France: Roles of Social- Ecological Legacies, Domestication, and Innovations
Copyright © 2012 by the author(s). Published here under license by the Resilience Alliance. Aumeeruddy-Thomas, Y., C. Therville, C. Lemarchand, A. Lauriac, and F. Richard. 2012. Resilience of sweet chestnut and truffle holm-oak rural forests in Languedoc-Roussillon, France: roles of social- ecological legacies, domestication, and innovations. Ecology and Society 17(2): 12. http://dx.doi.org/10.5751/ES-04750-170212 Research, part of a Special Feature on Public policies and management of rural forests: lasting alliance or fool’s dialogue? Resilience of Sweet Chestnut and Truffle Holm-Oak Rural Forests in Languedoc-Roussillon, France: Roles of Social-Ecological Legacies, Domestication, and Innovations Yildiz Aumeeruddy-Thomas 1, Clara Therville 2, Cedric Lemarchand 3, Alban Lauriac 4, and Franck Richard 1 ABSTRACT. The Cévennes sweet chestnut (Castanea sativa Mill.) forest-orchards and the holm-oak (Quercus ilex L.) black truffle (Tuber melanosporum Vittad.) associations of the garrigue in Languedoc-Roussillon have suffered a century of decline because of great reductions of rural populations and lack of understanding of the ecological and social dimensions of these rural forests by sectorial public agencies. Levels of tree and forest domestication alternated during historical periods in parallel with statuses of disorganization and reorganization of local social groups. Social-ecological legacies intrinsically linked to trees, forests, and landscape domestication, as well as knowledge, social, and technical practices have been mobilized and provided a basis for knowledge innovations, new domestications, uses, and new institutional networks related to changes in social set- ups. Collective actions emerging from local needs to revive territories in a modern context, cross-scale and reciprocal exchanges of rural and scientific knowledge, as well as institutional changes are interrelated variables that have enabled innovations and have increased resilience of these rural forests. -

Heritage Tree Walk
Trail Using the trail Refer to the map with trail markings inside this leaflet. A green plaque at each of the 12 trees along the trail will tell you the tree’s trail number, common name and botanical or Latin name. Stop at each tree and have a go at some of the activities. 1 Anna Nikiticheva 2 Wendy Caldon The other trails There are two other trails in the park. The Local people entered an art competition in 2013, Trees of the World trail in the West Park submitting pieces focussed Victoria Park highlights the variation of trees from around on the trees in the park. the world and their many uses throughout The competition winners were: history. In the East Park is a compact 1st prize - Anna Nikiticheva 2nd prize - Wendy Caldon Children’s Tree Walk around the play area 3rd prize - Radka Bailey near The Hub that includes a number of fun 3 Radka Bailey activities for children. Pick up your leaflets for all the trails at the Pavilion Cafe in the West Park or The Hub in the East Park. Heritage Tree Walk Trees of the World Children’s Tree Walk www.towerhamlets.gov.uk/victoriapark www.treesforcities.org, Registered Charity no. 1032154 Copper beech 4 Monkey puzzle/Chilean pine 7 11 Silver birch Welcome Tree info Araucara araucana Fagus sylvatica purpurea Betula pendula An evergreen tree growing to 40m Beech, like hazel, has been used for Silver birch provides food and habitat This is one of three tree trails created by tall. It can possibly live as long as centuries for its wood, specifically in for more than 300 insect species - Trees for Cities in partnership with Tower 1,000 years. -

Quercus Ilex Quercus Ilex, the Evergreen Oak, Holly Oak Or Holm Oak, Is a Large Evergreen Oak Native to the Mediterranean Region
Quercus ilex Quercus ilex, the evergreen oak, holly oak or holm oak, is a large evergreen oak native to the Mediterranean region. It takes its name from holm, an ancient name for holly. It is a member of the white oak section of the genus, with acorns that mature in a single summer. The first trees to be grown from acorns in England are still to be found within the stately grounds of Mamhead Park, Devon. From Britton & Brayley The Beauties of England and Wales (1803): Etymology The resemblance of the foliage to that of the common European holly Ilex aquifolium, has led to its common and botanic names. The name ilex was originally the classical Latin name for the holm oak, but later adopted as a botanical genus name for the hollies. Description An evergreen tree of large size, attaining in favourable places a height of 21–28 m, and developing in open situations a huge head of densely leafy branches as much across, the terminal portions of the branches usually pendulous in old trees. The trunk is sometimes over 6 m in girth. The young shoots are clothed with a close grey felt. The leaves are very variable in shape, most frequently narrowly oval or ovate-lanceolate, 4–8 cm long, 1.2–2.5 cm wide, rounded or broadly tapered at the base, pointed, the margins sometimes entire, sometimes (especially on young trees) more or less remotely toothed. When quite young, both surfaces are clothed with whitish down, which soon falls away entirely from the upper surface leaving it a dark glossy green; on the lower surface it turns grey or tawny, and persists until the fall of the leaf; the petiole is 3–16 mm long. -

Growth and Nutrient Acclimation of Evergreen Oak Seedlings Infected
Article Growth and Nutrient Acclimation of Evergreen Oak Seedlings Infected with Boletus reticulatus in Infertile Colluvial Soil in Warm Temperate Monsoon Asia: Evaluation of Early Growth Masazumi Kayama y Kyushu Research Center, Forestry and Forest Products Research Institute, Kumamoto 860-0862, Japan; kayama@affrc.go.jp; Tel.: +81-29-829-8219 Present address: Forestry and Forest Products Research Institute, Ibaraki 305-8687, Japan. y Received: 14 July 2020; Accepted: 6 August 2020; Published: 10 August 2020 Abstract: Soil erosion after harvesting of forest plantations can create infertile colluvial soil, therefore, seedlings used for site reforestation should be equipped against nutrient-poor edaphic conditions. The oak genus is a suitable candidate for such reforestation efforts. Oak is an ectomycorrhizal (ECM) tree genus known to grow under infertile environments. In this study, the initial stage of tree growth in three species of oak seedlings inoculated with a spore suspension of ECM fungus was monitored to evaluate the acceleration of seedling growth and nutrient uptake. I selected Quercus acuta Thunb., Quercus glauca Thunb., and Quercus salicina Blume, as these are common, evergreen, broad-leaved woody species commonly found in Southwestern Japan. The seedlings were inoculated with Boletus reticulatus and planted in infertile colluvial soils collected from a site that had undergone soil erosion. I also compared the ecophysiological characteristics of the potted seedlings planted in colluvial soil and normal forest soil. After six months of cultivation, Q. glauca with the ECM showed the highest growth rate in the fertile forest soil and had leaves with a higher nutrient content. In contrast, root dry mass increased slightly in Q. -

Population Genetic Diversity of Quercus Ilex Subsp. Ballota (Desf.) Samp
Article Population Genetic Diversity of Quercus ilex subsp. ballota (Desf.) Samp. Reveals Divergence in Recent and Evolutionary Migration Rates in the Spanish Dehesas Angel Fernández i Marti 1, Cristina Romero-Rodríguez 2, Rafael M. Navarro-Cerrillo 3 ID , Nieves Abril 2 ID , Jesús V. Jorrín-Novo 3 and Richard S. Dodd 1,* 1 Department of Environmental Science, Policy and Management, University of California Berkeley, Berkeley, CA 94720, USA; [email protected] 2 Agroforestry and Plant Biochemistry, Proteomics and Systems Biology, 14014 Cordoba, Spain; [email protected] (C.R.-R.); [email protected] (N.A.) 3 Evaluation and Restoration of Agricultural and Forestry Systems, Research Groups, University of Cordoba, UCO-CeiA3, 14014 Córdoba, Spain; [email protected] (R.M.N.-C.); [email protected] (J.V.J.-N.) * Correspondence: [email protected] Received: 3 April 2018; Accepted: 4 June 2018; Published: 7 June 2018 Abstract: The Spanish dehesas have been severely affected by human activities that date to the prehistoric period and have suffered accelerated decline since the 1980s. Holm oak (Quercus ilex subsp. ballota (Desf.) Samp.) is a key component of this system, and its acorns provide an important food source for wildlife and domesticated livestock. Our earlier work showed structured variation in acorn morphology and biochemistry. Here, we used chloroplast and nuclear microsatellites to detect genetic structure among populations of Q. ilex from the major biogeographic regions of Andalusia. We found high levels of spatial differentiation with chloroplast DNA indicating little seed dispersal among populations. Spatial differentiation was weaker for nuclear DNA, presumably as a result of more widespread pollen dispersal and its larger effective population size.