Fig S1 Genotyping
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MAP4K Family Kinases Act in Parallel to MST1/2 to Activate LATS1/2 in the Hippo Pathway
ARTICLE Received 19 Jun 2015 | Accepted 13 Aug 2015 | Published 5 Oct 2015 DOI: 10.1038/ncomms9357 OPEN MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway Zhipeng Meng1, Toshiro Moroishi1, Violaine Mottier-Pavie2, Steven W. Plouffe1, Carsten G. Hansen1, Audrey W. Hong1, Hyun Woo Park1, Jung-Soon Mo1, Wenqi Lu1, Shicong Lu1, Fabian Flores1, Fa-Xing Yu3, Georg Halder2 & Kun-Liang Guan1 The Hippo pathway plays a central role in tissue homoeostasis, and its dysregulation contributes to tumorigenesis. Core components of the Hippo pathway include a kinase cascade of MST1/2 and LATS1/2 and the transcription co-activators YAP/TAZ. In response to stimulation, LATS1/2 phosphorylate and inhibit YAP/TAZ, the main effectors of the Hippo pathway. Accumulating evidence suggests that MST1/2 are not required for the regulation of YAP/TAZ. Here we show that deletion of LATS1/2 but not MST1/2 abolishes YAP/TAZ phosphorylation. We have identified MAP4K family members—Drosophila Happyhour homologues MAP4K1/2/3 and Misshapen homologues MAP4K4/6/7—as direct LATS1/2-activating kinases. Combined deletion of MAP4Ks and MST1/2, but neither alone, suppresses phosphorylation of LATS1/2 and YAP/TAZ in response to a wide range of signals. Our results demonstrate that MAP4Ks act in parallel to and are partially redundant with MST1/2 in the regulation of LATS1/2 and YAP/TAZ, and establish MAP4Ks as components of the expanded Hippo pathway. 1 Department of Pharmacology and Moores Cancer Center, University of California San Diego, La Jolla, California 92093, USA. -

Biallelic Alteration and Dysregulation of the Hippo Pathway in Mucinous Tubular and Spindle Cell Carcinoma of the Kidney
Published OnlineFirst September 7, 2016; DOI: 10.1158/2159-8290.CD-16-0267 RESEARCH BRIEF Biallelic Alteration and Dysregulation of the Hippo Pathway in Mucinous Tubular and Spindle Cell Carcinoma of the Kidney Rohit Mehra 1 , 2 , 3 , Pankaj Vats 1 , 3 , 4 , Marcin Cieslik 3 , Xuhong Cao 3 , 5 , Fengyun Su 3 , Sudhanshu Shukla 3 , Aaron M. Udager 1 , Rui Wang 3 , Jincheng Pan 6 , Katayoon Kasaian 3 , Robert Lonigro 3 , Javed Siddiqui 3 , Kumpati Premkumar 4 , Ganesh Palapattu 7 , Alon Weizer 2 , 7 , Khaled S. Hafez 7 , J. Stuart Wolf Jr 7 , Ankur R. Sangoi 8 , Kiril Trpkov 9 , Adeboye O. Osunkoya 10 , Ming Zhou 11 , Giovanna A. Giannico 12 , Jesse K. McKenney 13 , Saravana M. Dhanasekaran 1 , 3 , and Arul M. Chinnaiyan 1 , 2 , 3 , 5 , 7 ABSTRACT Mucinous tubular and spindle cell carcinoma (MTSCC) is a relatively rare subtype of renal cell carcinoma (RCC) with distinctive morphologic and cytogenetic fea- tures. Here, we carry out whole-exome and transcriptome sequencing of a multi-institutional cohort of MTSCC (n = 22). We demonstrate the presence of either biallelic loss of Hippo pathway tumor sup- pressor genes (TSG) and/or evidence of alteration of Hippo pathway genes in 85% of samples. PTPN14 (31%) and NF2 (22%) were the most commonly implicated Hippo pathway genes, whereas other genes such as SAV1 and HIPK2 were also involved in a mutually exclusive fashion. Mutations in the context of recurrent chromosomal losses amounted to biallelic alterations in these TSGs. As a readout of Hippo pathway inactivation, a majority of cases (90%) exhibited increased nuclear YAP1 protein expression. -

DLG5 Connects Cell Polarity and Hippo Signaling Protein Networks by Linking PAR-1 with MST1/2
Downloaded from genesdev.cshlp.org on September 27, 2021 - Published by Cold Spring Harbor Laboratory Press DLG5 connects cell polarity and Hippo signaling protein networks by linking PAR-1 with MST1/2 Julian Kwan,1,2,7 Anna Sczaniecka,3,7 Emad Heidary Arash,1,4 Liem Nguyen,3 Chia-Chun Chen,3 Srdjana Ratkovic,2,5 Olga Klezovitch,3 Liliana Attisano,1,4 Helen McNeill,2,5 Andrew Emili,1,2 and Valeri Vasioukhin3,6 1Donnelly Centre for Cellular and Biomolecular Research, 2Department of Molecular Genetics, University of Toronto, Toronto, Ontario M5S 3E1, Canada; 3Division of Human Biology, Fred Hutchinson Cancer Research Center, Seattle, Washington 98109, USA; 4Department of Biochemistry, University of Toronto, Toronto, Ontario M5S 1A8, Canada; 5Lunenfeld-Tanenbaum Research Institute, Mt. Sinai Hospital, Toronto, Ontario M5G 1X5, Canada; 6Department of Pathology, Institute for Stem Cell and Regenerative Medicine, University of Washington, Seattle, Washington 98195, USA Disruption of apical–basal polarity is implicated in developmental disorders and cancer; however, the mechanisms connecting cell polarity proteins with intracellular signaling pathways are largely unknown. We determined previously that membrane-associated guanylate kinase (MAGUK) protein discs large homolog 5 (DLG5) functions in cell polarity and regulates cellular proliferation and differentiation via undefined mechanisms. We report here that DLG5 functions as an evolutionarily conserved scaffold and negative regulator of Hippo signaling, which controls organ size through the modulation of cell proliferation and differentiation. Affinity purification/mass spectrometry revealed a critical role of DLG5 in the formation of protein assemblies containing core Hippo kinases mammalian ste20 homologs 1/2 (MST1/2) and Par-1 polarity proteins microtubule affinity-regulating kinases 1/2/3 (MARK1/2/3). -

SAV1 Promotes Hippo Kinase Activation Through Antagonizing the PP2A Phosphatase STRIPAK
RESEARCH ARTICLE SAV1 promotes Hippo kinase activation through antagonizing the PP2A phosphatase STRIPAK Sung Jun Bae1†, Lisheng Ni1†, Adam Osinski1, Diana R Tomchick2, Chad A Brautigam2,3, Xuelian Luo1,2* 1Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, United States; 2Department of Biophysics, University of Texas Southwestern Medical Center, Dallas, United States; 3Department of Microbiology, University of Texas Southwestern Medical Center, Dallas, United States Abstract The Hippo pathway controls tissue growth and homeostasis through a central MST- LATS kinase cascade. The scaffold protein SAV1 promotes the activation of this kinase cascade, but the molecular mechanisms remain unknown. Here, we discover SAV1-mediated inhibition of the PP2A complex STRIPAKSLMAP as a key mechanism of MST1/2 activation. SLMAP binding to autophosphorylated MST2 linker recruits STRIPAK and promotes PP2A-mediated dephosphorylation of MST2 at the activation loop. Our structural and biochemical studies reveal that SAV1 and MST2 heterodimerize through their SARAH domains. Two SAV1–MST2 heterodimers further dimerize through SAV1 WW domains to form a heterotetramer, in which MST2 undergoes trans-autophosphorylation. SAV1 directly binds to STRIPAK and inhibits its phosphatase activity, protecting MST2 activation-loop phosphorylation. Genetic ablation of SLMAP in human cells leads to spontaneous activation of the Hippo pathway and alleviates the need for SAV1 in Hippo signaling. Thus, SAV1 promotes Hippo activation through counteracting the STRIPAKSLMAP PP2A *For correspondence: phosphatase complex. [email protected] DOI: https://doi.org/10.7554/eLife.30278.001 †These authors contributed equally to this work Competing interests: The Introduction authors declare that no The balance between cell division and death maintains tissue homeostasis of multicellular organisms. -

Structural Basis for Autoactivation of Human Mst2 Kinase and Its Regulation by RASSF5
Structure Article Structural Basis for Autoactivation of Human Mst2 Kinase and Its Regulation by RASSF5 Lisheng Ni,1 Sheng Li,1,4 Jianzhong Yu,3 Jungki Min,1,5 Chad A. Brautigam,2 Diana R. Tomchick,2 Duojia Pan,3 and Xuelian Luo1,* 1Department of Pharmacology 2Department of Biophysics The University of Texas Southwestern Medical Center, 6001 Forest Park Road, Dallas, TX 75390, USA 3Department of Molecular Biology and Genetics, Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA 4Present address: Biologics Department, Shanghai ChemPartner Co. Ltd., Shanghai 201203, China 5Present address: Department of Biochemistry, Duke University School of Medicine, Durham, NC 27710, USA *Correspondence: [email protected] http://dx.doi.org/10.1016/j.str.2013.07.008 SUMMARY dor (Sav1), the NDR family kinases Lats1 and Lats2 (Lats1/2), and the adaptor protein Mob1. They form a central kinase The tumor-suppressive Hippo pathway controls cascade to transduce signals from cell-surface receptors tissue homeostasis through balancing cell prolifera- (Avruch et al., 2012; Hergovich, 2012). tion and apoptosis. Activation of the kinases Mst1 In the canonical Hippo kinase cascade, Mst1/2, in complex and Mst2 (Mst1/2) is a key upstream event in this with Sav1, phosphorylate and activate the Lats1/2-Mob1 com- pathway and remains poorly understood. Mst1/2 plexes, which then phosphorylate the transcriptional coactivator and their critical regulators RASSFs contain Yes-associated protein (YAP), a major downstream target of the Hippo pathway (Dong et al., 2007; Hao et al., 2008; Hong and Salvador/RASSF1A/Hippo (SARAH) domains that Guan, 2012; Huang et al., 2005; Zhao et al., 2007). -

The Role of the C-Terminus Merlin in Its Tumor Suppressor Function Vinay Mandati
The role of the C-terminus Merlin in its tumor suppressor function Vinay Mandati To cite this version: Vinay Mandati. The role of the C-terminus Merlin in its tumor suppressor function. Agricultural sciences. Université Paris Sud - Paris XI, 2013. English. NNT : 2013PA112140. tel-01124131 HAL Id: tel-01124131 https://tel.archives-ouvertes.fr/tel-01124131 Submitted on 19 Mar 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. 1 TABLE OF CONTENTS Abbreviations ……………………………………………………………………………...... 8 Resume …………………………………………………………………………………… 10 Abstract …………………………………………………………………………………….. 11 1. Introduction ………………………………………………………………………………12 1.1 Neurofibromatoses ……………………………………………………………………….14 1.2 NF2 disease ………………………………………………………………………………15 1.3 The NF2 gene …………………………………………………………………………….17 1.4 Mutational spectrum of NF2 gene ………………………………………………………..18 1.5 NF2 in other cancers ……………………………………………………………………...20 2. ERM proteins and Merlin ……………………………………………………………….21 2.1 ERMs ……………………………………………………………………………………..21 2.1.1 Band 4.1 Proteins and ERMs …………………………………………………………...21 2.1.2 ERMs structure ………………………………………………………………………....23 2.1.3 Sub-cellular localization and tissue distribution of ERMs ……………………………..25 2.1.4 ERM proteins and their binding partners ……………………………………………….25 2.1.5 Assimilation of ERMs into signaling pathways ………………………………………...26 2.1.5. A. ERMs and Ras signaling …………………………………………………...26 2.1.5. B. ERMs in membrane transport ………………………………………………29 2.1.6 ERM functions in metastasis …………………………………………………………...30 2.1.7 Regulation of ERM proteins activity …………………………………………………...31 2.1.7. -

SAV1 Monoclonal Antibody (M02), Clone 3B2
SAV1 monoclonal antibody (M02), clone 3B2 Catalog # : H00060485-M02 規格 : [ 100 ug ] List All Specification Application Image Product Mouse monoclonal antibody raised against a partial recombinant SAV1. Western Blot (Cell lysate) Description: Immunogen: SAV1 (NP_068590, 300 a.a. ~ 383 a.a) partial recombinant protein with GST tag. MW of the GST tag alone is 26 KDa. Sequence: HTAEIPDWLQVYARAPVKYDHILKWELFQLADLDTYQGMLKLLFMKELE QIVKMYEAYRQALLTELENRKQRQQWYAQQHGKNF enlarge Western Blot (Cell lysate) Host: Mouse Reactivity: Human Isotype: IgG2a Kappa Quality Control Antibody Reactive Against Recombinant Protein. enlarge Testing: Western Blot (Recombinant protein) Immunofluorescence enlarge Immunoprecipitation Western Blot detection against Immunogen (34.98 KDa) . Storage Buffer: In 1x PBS, pH 7.4 Storage Store at -20°C or lower. Aliquot to avoid repeated freezing and thawing. Instruction: enlarge MSDS: Download Sandwich ELISA (Recombinant protein) Datasheet: Download Publication Reference 1. Kidney-specific knockout of Sav1 in the mouse promotes hyper-proliferation of renal tubular epithelium through suppression of the Hippo pathway. Kai T, Tsukamoto Y, Hijiya N, Tokunaga A, Nakada C, Uchida T, Daa T, Iha H, enlarge Takahashi M, Nomura T, Sato F, Mimata H, Ikawa M, Seto M, Matsuura K, Moriyama M.J Pathol. 2016 Mar;239(1):97-108. ELISA 2. Hippo signaling mediates proliferation, invasiveness and metastatic potential of clear cell renal cell carcinoma. Schutte U, Bisht S, Heukamp LC, Kebschull M, Florin A, Haarmann J, Hoffmann P, Bendas G, Buettner R, Brossart P, Feldmann G.Translational Oncology Volume 7, Issue 2, April 2014, Pages 309–321 Page 1 of 4 2018/1/10 3. Screening of binding proteins that interact with human Salvador 1 in a human fetal liver cDNA library by the yeast two-hybrid system. -

The Hippo–YAP Pathway in Organ Size Control and Tumorigenesis: an Updated Version
Downloaded from genesdev.cshlp.org on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW The Hippo–YAP pathway in organ size control and tumorigenesis: an updated version Bin Zhao,1 Li Li,1 Qunying Lei,2 and Kun-Liang Guan1,3 1Department of Pharmacology and Moores Cancer Center, University of California at San Diego, La Jolla, California 92093, USA; 2Department of Biological Chemistry, School of Medicine, and Molecular and Cell Biology Laboratory, Institutes of Biomedical Sciences, Fudan University, Shanghai 200032, China The Hippo signaling pathway is gaining recognition as The Hippo pathway was named after the Drosophila an important player in both organ size control and Hippo kinase that was discovered using this approach. tumorigenesis, which are physiological and pathological Components of the Hippo pathway are highly conserved processes that share common cellular signaling mecha- in mammals (Fig. 1). Later genetic and biochemical studies nisms. Upon activation by stimuli such as high cell den- gradually shaped the current working model, in which the sity in cell culture, the Hippo pathway kinase cascade mammalian Mst1/2 kinase (Hippo homolog), complexed phosphorylates and inhibits the Yes-associated protein with a scaffold protein, Sav1, phosphorylates and activates (YAP)/TAZ transcription coactivators representing the the Lats1/2 kinase. Lats1/2 is also activated by another major signaling output of the pathway. Altered gene scaffold protein, Mob1 (Fig. 2). These four proteins are expression resulting from YAP/TAZ inhibition affects often referred to as the core components of the Hippo cell number by repressing cell proliferation and promot- pathway. At the upstream, several components have ing apoptosis, thereby limiting organ size. -
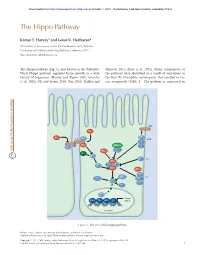
The Hippo Pathway
Downloaded from http://cshperspectives.cshlp.org/ on October 1, 2021 - Published by Cold Spring Harbor Laboratory Press The Hippo Pathway Kieran F. Harvey1 and Iswar K. Hariharan2 1Peter MacCallum Cancer Centre, East Melbourne 3002, Australia 2University of California, Berkeley, Berkeley, California 94720 Correspondence: [email protected] The Hippo pathway (Fig. 1), also known as the Salvador- Johnson 2011; Zhao et al. 2011). Many components of Warts-Hippo pathway, regulates tissue growth in a wide the pathway were identified as a result of mutations in variety of organisms (Harvey and Tapon 2007; Grusche the fruit fly Drosophila melanogaster that resulted in tis- et al. 2010; Oh and Irvine 2010; Pan 2010; Halder and sue overgrowth (Table 1). The pathway is conserved in CRB FJ FJ EX FT LFT SAR Kibra DCO STRIPAK LFT DS MER TAO1 RASSF APP D JUB AJ HPO ZYX MATS SAV WTS αPKC LGL 14-3-3 SJ WBP2 SCRIB YKI DLG MOP YKI Target MAD TSH HTH SD genes Nucleus Figure 1. The Drosophila Hippo pathway. Editors: Lewis Cantley, Tony Hunter, Richard Sever, and Jeremy W. Thorner Additional Perspectives on Signal Transduction available at www.cshperspectives.org Copyright # 2012 Cold Spring Harbor Laboratory Press; all rights reserved; doi: 10.1101/cshperspect.a011288 Cite this article as Cold Spring Harb Perspect Biol 2012;4:a011288 1 Downloaded from http://cshperspectives.cshlp.org/ on October 1, 2021 - Published by Cold Spring Harbor Laboratory Press K.F. Harvey and I.K. Hariharan Table 1. Components of the Drosophila melanogaster Salvador- Warts (WTS; also known as LATS) (Justice et al. -

The MST1/2-SAV1 Complex of the Hippo Pathway Promotes Ciliogenesis
ARTICLE Received 27 Mar 2014 | Accepted 25 Sep 2014 | Published 4 Nov 2014 DOI: 10.1038/ncomms6370 The MST1/2-SAV1 complex of the Hippo pathway promotes ciliogenesis Miju Kim1, Minchul Kim1, Mi-Sun Lee2, Cheol-Hee Kim2 & Dae-Sik Lim1 Primary cilia are microtubule-based organelles that protrude from polarized epithelial cells. Although many structural and trafficking molecules that regulate ciliogenesis have been discovered, signalling proteins are not well defined. Here we show that the MST1/2-SAV1 complex, a core component of the Hippo pathway, promotes ciliogenesis. MST1 is activated during ciliogenesis and localizes to the basal body of cilia. Depletion of MST1/2 or SAV1 impairs ciliogenesis in cultured cells and induces ciliopathy phenotypes in zebrafish. MST1/ 2-SAV1 regulates ciliogenesis through two independent mechanisms: MST1/2 binds and phosphorylates Aurora kinase A (AURKA), leading to dissociation of the AURKA/HDAC6 cilia-disassembly complex; and MST1/2-SAV1 associates with the NPHP transition-zone complex, promoting ciliary localization of multiple ciliary cargoes. Our results suggest that components of the Hippo pathway contribute to establish a polarized cell structure in addition to regulating proliferation. 1 Department of Biological Sciences, National Creative Research Initiatives Center, Korea Advanced Institute of Science and Technology (KAIST), Daejeon 305-701, Korea. 2 Department of Biology, Chungnam National University, Daejeon 305-764, Korea. Correspondence and requests for materials should be addressed to D.-S.L. (email: [email protected]). NATURE COMMUNICATIONS | 5:5370 | DOI: 10.1038/ncomms6370 | www.nature.com/naturecommunications 1 & 2014 Macmillan Publishers Limited. All rights reserved. ARTICLE NATURE COMMUNICATIONS | DOI: 10.1038/ncomms6370 rimary cilia are microtubule-based organelles that extrude dissociation of the AURKA/HDAC6 cilia-disassembly complex; from the apical surface of epithelial cells and function as and (2) MST1/2-SAV1 associates with the NPHP transition-zone Pantennae that monitor the extracellular environment1. -

LIM-Domain Protein AJUBA Suppresses Malignant Mesothelioma Cell Proliferation Via Hippo Signaling Cascade
Oncogene (2015) 34, 73–83 & 2015 Macmillan Publishers Limited All rights reserved 0950-9232/15 www.nature.com/onc ORIGINAL ARTICLE LIM-domain protein AJUBA suppresses malignant mesothelioma cell proliferation via Hippo signaling cascade I Tanaka1,2,6, H Osada1,3, M Fujii1, A Fukatsu1,3, T Hida4, Y Horio4, Y Kondo1, A Sato5, Y Hasegawa2, T Tsujimura5 and Y Sekido1,3 Malignant mesothelioma (MM) is one of the most aggressive neoplasms usually associated with asbestos exposure and is highly refractory to current therapeutic modalities. MMs show frequent activation of a transcriptional coactivator Yes-associated protein (YAP), which is attributed to the neurofibromatosis type 2 (NF2)–Hippo pathway dysfunction, leading to deregulated cell proliferation and acquisition of a malignant phenotype. However, the whole mechanism of disordered YAP activation in MMs has not yet been well clarified. In the present study, we investigated various components of the NF2-Hippo pathway, and eventually found that MM cells frequently showed downregulation of LIM-domain protein AJUBA, a binding partner of large tumor suppressor type 2 (LATS2), which is one of the last-step kinases of the NF2-Hippo pathway. Although loss of AJUBA expression was independent of the alteration status of other Hippo pathway components, MM cell lines with AJUBA inactivation showed a more dephosphorylated (activated) level of YAP. Immunohistochemical analysis showed frequent downregulation of AJUBA in primary MMs, which was associated with YAP constitutive activation. We found that AJUBA transduction into MM cells significantly suppressed promoter activities of YAP-target genes, and the suppression of YAP activity by AJUBA was remarkably canceled by knockdown of LATS2. -

The Hippo Signaling Pathway in Drug Resistance in Cancer
cancers Review The Hippo Signaling Pathway in Drug Resistance in Cancer Renya Zeng and Jixin Dong * Eppley Institute for Research in Cancer and Allied Diseases, Fred & Pamela Buffett Cancer Center, University of Nebraska Medical Center, Omaha, NE 68198, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-402-559-5596; Fax: +1-402-559-4651 Simple Summary: Although great breakthroughs have been made in cancer treatment following the development of targeted therapy and immune therapy, resistance against anti-cancer drugs remains one of the most challenging conundrums. Considerable effort has been made to discover the underlying mechanisms through which malignant tumor cells acquire or develop resistance to anti-cancer treatment. The Hippo signaling pathway appears to play an important role in this process. This review focuses on how components in the human Hippo signaling pathway contribute to drug resistance in a variety of cancer types. This article also summarizes current pharmacological interventions that are able to target the Hippo signaling pathway and serve as potential anti-cancer therapeutics. Abstract: Chemotherapy represents one of the most efficacious strategies to treat cancer patients, bringing advantageous changes at least temporarily even to those patients with incurable malignan- cies. However, most patients respond poorly after a certain number of cycles of treatment due to the development of drug resistance. Resistance to drugs administrated to cancer patients greatly limits the benefits that patients can achieve and continues to be a severe clinical difficulty. Among the mechanisms which have been uncovered to mediate anti-cancer drug resistance, the Hippo signaling pathway is gaining increasing attention due to the remarkable oncogenic activities of its components (for example, YAP and TAZ) and their druggable properties.