A Maximum Subsurface Biomass on Mars from Untapped Free Energy: CO
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Stringent Upper Limit of 20 Pptv for Methane on Mars and Constraints on Its Dispersion Outside Gale Crater F
A&A 650, A140 (2021) Astronomy https://doi.org/10.1051/0004-6361/202140389 & © F. Montmessin et al. 2021 Astrophysics A stringent upper limit of 20 pptv for methane on Mars and constraints on its dispersion outside Gale crater F. Montmessin1 , O. I. Korablev2 , A. Trokhimovskiy2 , F. Lefèvre3 , A. A. Fedorova2 , L. Baggio1 , A. Irbah1 , G. Lacombe1, K. S. Olsen4 , A. S. Braude1 , D. A. Belyaev2 , J. Alday4 , F. Forget5 , F. Daerden6, J. Pla-Garcia7,8 , S. Rafkin8 , C. F. Wilson4, A. Patrakeev2, A. Shakun2, and J. L. Bertaux1 1 LATMOS/IPSL, UVSQ Université Paris-Saclay, Sorbonne Université, CNRS, Guyancourt, France e-mail: [email protected] 2 Space Research Institute (IKI) RAS, Moscow, Russia 3 LATMOS/IPSL, Sorbonne Université, UVSQ Université Paris-Saclay, CNRS, Paris, France 4 AOPP, Oxford University, Oxford, UK 5 Laboratoire de Météorologie Dynamique, Sorbonne Université, Paris, France 6 BIRA-IASB, Bruxelles, Belgium 7 Centro de Astrobiología (CSIC?INTA), Torrejón de Ardoz, Spain 8 Southwest Research Institute, Boulder, CO, USA Received 20 January 2021 / Accepted 23 April 2021 ABSTRACT Context. Reports on the detection of methane in the Martian atmosphere have motivated numerous studies aiming to confirm or explain its presence on a planet where it might imply a biogenic or more likely a geophysical origin. Aims. Our intent is to complement and improve on the previously reported detection attempts by the Atmospheric Chemistry Suite (ACS) on board the ExoMars Trace Gas Orbiter (TGO). This latter study reported the results of a campaign that was a few months in length, and was significantly hindered by a dusty period that impaired detection performances. -

The Mystery of Methane on Mars and Titan
The Mystery of Methane on Mars & Titan By Sushil K. Atreya MARS has long been thought of as a possible abode of life. The discovery of methane in its atmosphere has rekindled those visions. The visible face of Mars looks nearly static, apart from a few wispy clouds (white). But the methane hints at a beehive of biological or geochemical activity underground. Of all the planets in the solar system other than Earth, own way, revealing either that we are not alone in the universe Mars has arguably the greatest potential for life, either extinct or that both Mars and Titan harbor large underground bodies or extant. It resembles Earth in so many ways: its formation of water together with unexpected levels of geochemical activ- process, its early climate history, its reservoirs of water, its vol- ity. Understanding the origin and fate of methane on these bod- canoes and other geologic processes. Microorganisms would fit ies will provide crucial clues to the processes that shape the right in. Another planetary body, Saturn’s largest moon Titan, formation, evolution and habitability of terrestrial worlds in also routinely comes up in discussions of extraterrestrial biology. this solar system and possibly in others. In its primordial past, Titan possessed conditions conducive to Methane (CH4) is abundant on the giant planets—Jupiter, the formation of molecular precursors of life, and some scientists Saturn, Uranus and Neptune—where it was the product of chem- believe it may have been alive then and might even be alive now. ical processing of primordial solar nebula material. On Earth, To add intrigue to these possibilities, astronomers studying though, methane is special. -

Evidence for Methane in Martian Meteorites
Blamey, N. J. F., Parnell, J., McMahon, S., Mark, D. F., Tomkinson, T., Lee, M., Shivak, J., Izawa, M. R. M., Banerjee, N. R., & Flemming, R. L. (2015). Evidence for methane in Martian meteorites. Nature Communications, 6, [7399]. https://doi.org/10.1038/ncomms8399 Publisher's PDF, also known as Version of record License (if available): CC BY Link to published version (if available): 10.1038/ncomms8399 Link to publication record in Explore Bristol Research PDF-document This is the final published version of the article (version of record). It first appeared online via Nature Publishing Group at doi:10.1038/ncomms8399. Please refer to any applicable terms of use of the publisher. University of Bristol - Explore Bristol Research General rights This document is made available in accordance with publisher policies. Please cite only the published version using the reference above. Full terms of use are available: http://www.bristol.ac.uk/red/research-policy/pure/user-guides/ebr-terms/ ARTICLE Received 19 Sep 2014 | Accepted 1 May 2015 | Published 16 Jun 2015 DOI: 10.1038/ncomms8399 OPEN Evidence for methane in Martian meteorites Nigel J.F. Blamey1,2, John Parnell2, Sean McMahon2, Darren F. Mark3, Tim Tomkinson3,4, Martin Lee4, Jared Shivak5, Matthew R.M. Izawa5, Neil R. Banerjee5 & Roberta L. Flemming5 The putative occurrence of methane in the Martian atmosphere has had a major influence on the exploration of Mars, especially by the implication of active biology. The occurrence has not been borne out by measurements of atmosphere by the MSL rover Curiosity but, as on Earth, methane on Mars is most likely in the subsurface of the crust. -

Life on Mars- Extracting the Signs for Future Human Habitation on The
Muhammad Shadab Khan, Department of Aeronautical Engineering, Babu Banarasi Das National Institute of Technology and Management, Lucknow, India CONTENT- Introduction Life on Mars Signs of Life Found on Mars and Similarities with those signs on Earth Utilizing Life Support Resources present on Mars for human habitation INTRODUCTION 1- Rise of Global Warming and it’s alarming consequences which might cause complete loss of life on Earth– Search for an Earth like habitable body in the outer Space 2- Ongoing Exploration of Mars in search of Life on the Red Planet for future human habitation on the planet. 3- Mars is the planet which is very similar to Earth and it’s believed that Life had existed on Mars in the Past– Big support to our quest for life in the future 4- The life supporting features found on Mars are very similar to those found on Earth– Giving a sign of prospective life in the near future on Mars Life on Mars Life on Mars has been a subject of wide discussion from the very early beginning. There has been speculations about the existence of Life on the Red Planet in the Past But still about the presence of an Earth like habitable environment on Mars is a BIG QUESTION ? The first search for Life on Mars was carried out by NASA’s VIKING LANDER in 1976 Gas Chromatograph/Mass Spectrometer Instrument on the Lander detected the possible MICROBIAL LIFE in the soil sample . The question about MICROBIAL LIFE remains unresolved LIQUID WATER ON MARS The speculation about the presence of LIQUID WATER on Mars has been a subject of deep interest to the scientists from the very beginning As it’s believed in order to support LIFE, the need of WATER in LIQUID FORM is MUST. -

Schultze-Makuch Methane Mars.Pdf
International Journal of Astrobiology 7 (2): 117–141 (2008) Printed in the United Kingdom 117 doi:10.1017/S1473550408004175 f 2008 Cambridge University Press The case for life on Mars Dirk Schulze-Makuch1, Alberto G. Faire´n2 and Alfonso F. Davila2 1School of Earth and Environmental Sciences, Washington State University, Pullman, WA 99163, USA e-mail: [email protected] 2Space Science and Astrobiology Division, NASA Ames Research Center, USA Abstract: There have been several attempts to answer the question of whether there is, or has ever been, life on Mars. The boldest attempt was the only ever life detection experiment conducted on another planet: the Viking mission. The mission was a great success, but it failed to provide a clear answer to the question of life on Mars. More than 30 years after the Viking mission our understanding of the history and evolution of Mars has increased vastly to reveal a wetter Martian past and the occurrence of diverse environments that could have supported microbial life similar to that on Earth for extended periods of time. The discovery of Terran extremophilic microorganisms, adapted to environments previously though to be prohibitive for life, has greatly expanded the limits of habitability in our Solar System, and has opened new avenues for the search of life on Mars. Remnants of a possible early biosphere may be found in the Martian meteorite ALH84001. This claim is based on a collection of facts and observations consistent with biogenic origins, but individual links in the collective chain of evidence remain controversial. Recent evidence for contemporary liquid water on Mars and the detection of methane in the Martian atmosphere further enhance the case for life on Mars. -

Methane on Mars: New Insights Into the Sensitivity of CH4 with the NOMAD/Exomars Spectrometer Through Its First In-Flight Calibration
Methane on Mars: new insights into the sensitivity of CH4 with the NOMAD/ExoMars spectrometer through its first in-flight calibration Giuliano Liuzzia,b, Geronimo L. Villanuevaa, Michael J. Mummaa, Michael D. Smitha, Frank Daerdenc, Bojan Risticc, Ian Thomasc, Ann Carine Vandaelec, Manish R. Pateld,e, José-Juan Lopez-Morenof, Giancarlo Belluccig, and the NOMAD team a NASA Goddard Space Flight Center, 8800 Greenbelt Rd., Greenbelt, 20771 MD, USA b Dep. of Physics, American University, 4400 Massachusetts Av., Washington, 20016 DC, USA c Belgian Institute for Space Aeronomy, BIRA-IASB, Ringlaan 3, 1180 Brussels, Belgium d School of Physical Sciences, The Open University, Milton Keynes, MK7 6AA, UK e SSTD, STFC Rutherford Appleton Laboratory, Chilton, Oxfordshire OX11 0QX, UK f Instituto de Astrofisica de Andalucia, IAA-CSIC, Glorieta de la Astronomia, 18008 Granada, Spain g Istituto di Astrofisica e Planetologia Spaziali, IAPS-INAF, Via del Fosso del Cavaliere 100, 00133 Rome, Italy Corresponding author: Giuliano Liuzzi, e-mail: [email protected] Abstract The Nadir and Occultation for MArs Discovery instrument (NOMAD), onboard the ExoMars Trace Gas Orbiter (TGO) spacecraft was conceived to observe Mars in solar occultation, nadir, and limb geometries, and will be able to produce an outstanding amount of diverse data, mostly focused on properties of the atmosphere. The infrared channels of the instrument operate by combining an echelle grating spectrometer with an Acousto-Optical Tunable Filter (AOTF). Using in-flight data, we characterized the instrument performance and parameterized its calibration. In particular: an accurate frequency calibration was achieved, together with its variability due to thermal effects on the grating. -

Critically Testing Olivine-Hosted Putative Martian Biosignatures in the Yamato 000593
1 Critically testing olivine-hosted putative Martian biosignatures in the Yamato 000593 2 meteorite - geobiological implications 3 4 Abstract: 5 On rocky planets such as Earth and Mars the serpentinization of olivine in ultramafic crust 6 produces hydrogen that can act as a potential energy source for life. Direct evidence of fluid-rock 7 interaction on Mars comes from iddingsite alteration veins found in Martian meteorites. In the 8 Yamato 000593 meteorite putative biosignatures have been reported from altered olivines in the 9 form of microtextures and associated organic material that have been compared to tubular 10 bioalteration textures found in terrestrial sub-seafloor volcanic rocks. Here we use a suite of 11 correlative, high-sensitivity, in-situ chemical and morphological analyses to characterize and re- 12 evaluate these microalteration textures in Yamato 000593, a clinopyroxenite from the shallow sub- 13 surface of Mars. We show that the altered olivine crystals have angular and micro-brecciated 14 margins and are also highly strained due to impact induced fracturing. The shape of the olivine 15 microalteration textures is in no way comparable to microtunnels of inferred biological origin 16 found in terrestrial volcanic glasses and dunites, and rather we argue that the Yamato 000593 17 microtextures are abiotic in origin. Vein filling iddingsite extends into the olivine microalteration 18 textures and contains amorphous organic carbon occurring as bands and sub-spherical 19 concentrations <300 nm across. We propose that a Martian impact event produced the micro- 20 brecciated olivine crystal margins that reacted with subsurface hydrothermal fluids to form 21 iddingsite containing organic carbon derived from abiotic sources. -
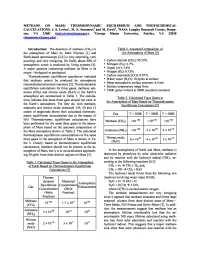
Methane on Mars: Thermodynamic Equilibrium and Photochemical Calculations
METHANE ON MARS: THERMODYNAMIC EQUILIBRIUM AND PHOTOCHEMICAL CALCULATIONS. J. S. Levine', M. E. Summers2 and M. Ewell2, 'NASA Langley Research Center, Hamp- ton, VA 2368' ([email protected] ), 2Geroge Mason University, Fairfax, VA 22030 ([email protected]) . Introduction: The detection of methane (CH4) in Table 2. Assumed Composition of the atmosphere of Mars by Mars Express [1] and the Atmosphere of Mars [ 5] Earth-based spectroscopy [2,3] is very surprising, very puzzling, and very intriguing. On Earth, about 90% of • Carbon dioxide (CO 2): 95.32% atmospheric ozone is produced by living systems [4]. • Nitrogen (N 2): 2.7% A major question concerning methane on Mars is its • Argon (Ar): 1.6% origin —biological or geological. • Oxygen (O 2): 0.13% Thermodynamic equilibrium caculations indicated • Carbon monoxide (CO): 0.07% that methane cannot be produced by atmospheric • Water vapor (H 2O): 10 ppmv at surface • Mean atmospheric surface pressure: 6.4 mb chemical/photochemical reactions [5]. Thermodynamic • Surface temperature range from equilibrium calculations for three gases, methane, am- • 148K (polar winter) to 290K (southern summer) monia (NH3) and nitrous oxide (N 2 O) in the Earth’s atmosphere are summarized in Table 1. The calcula- Table 3. Calculated Trace Gases in tions indicate that these three gses should not exist in the Atmosphere of Mars Based on Thermodynamic the Earth’s atmosphere. Yet they do, with methane, Equilibrium Calculations [5] ammonia and nitrous oxide enhanced 139, 50 and 12 orders of magnitude above their calculated thermody- Gas T=100KT= 200K T = 300K namic equilibrium concentration due to the impact of life! Thermodynamic equilibrium calculations have -100 -100 -100 Methane (CH4) <10 <10 <10 been performed for the same three gases in the atmos- phere of Mars based on the assumed composition of -100 -89 -62 the Mars atmosphere shown in Table 2. -

The Search and Prospects for Life on Mars Overview
AST 309 part 2: Extraterrestrial Life The Search and Prospects for Life on Mars Overview: • Life on Mars: a historical view • Present-day Mars • The Exploration of Mars • The Viking Mission • AHL 84001 • Mars Exploration Rovers • Evidence for water • Methane on Mars! • The future Our perception of Mars through history: • named after the roman god of war (probably due to its color) • (mainly) used by Johannes Kepler to derive laws of planetary motion • early observations with telescopes showed polar caps, dark areas and moons => very Earth-like? • Percival Lowell: canals on Mars? Intelligent life? Lowell in the 1890s Our perception of Mars through history: H.G. Wells: “The War of the Worlds” (1898) Orson Wells’ radio broadcast in 1938 Modern exploration of Mars: Mariner 4 Early missions: 1964 Mariner 4 first flyby 1971 Mariner 9 orbits Mars Mars 3 & 4 land (but stopped working) 1976 2 Viking orbiter + Landers 1988 Phobos 1 & 2 failed 1992 Mars Observer failed 1997 Mars Global Surveyor & Mars Pathfinder begin modern era Olympus Mons Mars basic facts: Distance: 1.5 AU Period: 1.87 years Radius: 0.53 R_Earth Mass: 0.11 M_Earth Density: 4.0 g/cm3 Satellites: Phobos and Deimos Structure: Dense Core (~1700 km), rocky mantle, thin crust Temperature: -87 to -5 C Magnetic Field: Weak and variable (some parts strong) Atmosphere: 95% CO2, 3% Nitrogen, argon, traces of oxygen Mars atmosphere: Very thin! ~0.7% of Earth’s pressure Pressure can change significantly because it gets so cold that CO2 freezes out on polar caps Viking atmospheric measurements 95.3% carbon dioxide 2.7% nitrogen 1.6% argon 0.13% oxygen Composition 0.07% carbon monoxide 0.03% water vapor trace neon, krypton, xenon, ozone, methane 1-9 millibars, depending on Surface pressure altitude; average 7 mb Viking on Mars: Viking 1 Lander touched down at Chryse Planitia (22.48° N, 49.97° W planetographic, 1.5 km below the datum (6.1 mbar) elevation). -

Chemical Methods for Searching for Evidence of Extra-Terrestrial Life
Downloaded from http://rsta.royalsocietypublishing.org/ on June 27, 2016 Phil. Trans. R. Soc. A (2011) 369, 607–619 doi:10.1098/rsta.2010.0241 REVIEW Chemical methods for searching for evidence of extra-terrestrial life BY COLIN PILLINGER* Planetary and Space Sciences Research Institute, The Open University, Walton Hall, Milton Keynes MK7 6AA, UK This paper describes the chemical concepts used for the purpose of detecting life in extra-terrestrial situations. These methods, developed initially within the oil industry, have been used to determine when life began on Earth and for investigating the Moon and Mars via space missions. In the case of Mars, the Viking missions led to the realization that we had meteorites from Mars on Earth. The study of Martian meteorites in the laboratory provides tantalizing clues for life on Mars in both the ancient and recent past. Meteorite analyses led to the launch of the Beagle 2 spacecraft, which was designed to prove that life-detection results obtained on Earth were authentic and not confused by terrestrial contamination. Some suggestions are made for future work. Keywords: biomarker; carbon skeleton; isotope 1. Historical studies There are a variety of concepts involved in using chemical methods to detect evidence of life during laboratory studies of samples on Earth or via instrumentation on planetary lander spacecraft. They comprise indentifying exact chemical structure, measuring precise stable isotope composition or a mixture of both. 2. Introduction Perhaps, the second most important molecule concerning living things on Earth is (i) chlorophyll-a. It fulfils the function of translating the energy carried by sunlight into chemical energy stored as carbohydrates. -

Methane Release on Early Mars by Atmospheric Collapse and Atmospheric Reinflation
Methane release on Early Mars by atmospheric collapse and atmospheric reinflation Edwin S. Kite1, Michael A. Mischna2, Peter Gao3,4, Yuk L. Yung2,5, Martin Turbet6,7,* 1. University of Chicago ([email protected]), Chicago, IL 60637. 2. Jet Propulsion Laboratory, California Institute of Technology, Pasadena, CA 91109. 3. NASA Ames Research Center, Mountain View, CA 94035. 4. University of California, Berkeley, CA 94720. 5. California Institute of Technology, Pasadena, CA 91125. 6. Laboratoire de Météorologie Dynamique, IPSL, Sorbonne Universités, UPMC Univ Paris 06, CNRS, Paris 75005, France. 7. Observatoire Astronomique de l’Université de Genève, 51 Chemin des Maillettes, 1290 Sauverny, Switzerland. * Currently Marie Sklodowska-Curie Fellow, Geneva Astronomical Observatory. Abstract. A candidate explanation for Early Mars rivers is atmospheric warming due to surface release of H2 or CH4 gas. However, it remains unknown how much gas could be released in a single event. We model the CH4 release by one mechanism for rapid release of CH4 from clathrate. By modeling how CH4-clathrate release is affected by changes in Mars’ obliquity and atmospheric composition, we find that a large fraction of total outgassing from CH4 clathrate occurs following Mars’ first prolonged atmospheric collapse. This atmosphere-collapse-initiated CH4-release mechanism has three stages. (1) Rapid collapse of Early Mars’ carbon dioxide atmosphere initiates a slower shift of water ice from high ground to the poles. (2) Upon subsequent CO2-atmosphere re-inflation and CO2-greenhouse warming, low-latitude clathrate decomposes and releases methane gas. (3) Methane can then perturb atmospheric chemistry and surface temperature, until photochemical processes destroy the methane. -

Evidence for Methane in Martian Meteorites
ARTICLE Received 19 Sep 2014 | Accepted 1 May 2015 | Published 16 Jun 2015 DOI: 10.1038/ncomms8399 Evidence for methane in Martian meteorites Nigel J.F. Blamey1,2, John Parnell2, Sean McMahon2, Darren F. Mark3, Tim Tomkinson3,4, Martin Lee4, Jared Shivak5, Matthew R.M. Izawa5, Neil R. Banerjee5 & Roberta L. Flemming5 The putative occurrence of methane in the Martian atmosphere has had a major influence on the exploration of Mars, especially by the implication of active biology. The occurrence has not been borne out by measurements of atmosphere by the MSL rover Curiosity but, as on Earth, methane on Mars is most likely in the subsurface of the crust. Serpentinization of olivine-bearing rocks, to yield hydrogen that may further react with carbon-bearing species, has been widely invoked as a source of methane on Mars, but this possibility has not hitherto been tested. Here we show that some Martian meteorites, representing basic igneous rocks, liberate a methane-rich volatile component on crushing. The occurrence of methane in Martian rock samples adds strong weight to models whereby any life on Mars is/was likely to be resident in a subsurface habitat, where methane could be a source of energy and carbon for microbial activity. 1 Department of Earth Sciences, Brock University, 500 Glenridge Avenue, St Catharines, Ontario, Canada L2S 3A1. 2 School of Geosciences, University of Aberdeen, Aberdeen AB24 3UE, UK. 3 Scottish Universities Environmental Research Centre, Glasgow G75 0QF, UK. 4 School of Geographical and Earth Sciences, University of Glasgow, Glasgow G12 8QQ, UK. 5 Department of Earth Sciences, University of Western Ontario, Ontario, Canada N6A 5B7.