Posttranslational Modifications of RUNX1 As Potential Anticancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MECOM Permits Pancreatic Acinar Cell Dedifferentiation Avoiding Cell Death Under Stress Conditions
Cell Death & Differentiation (2021) 28:2601–2615 https://doi.org/10.1038/s41418-021-00771-6 ARTICLE MECOM permits pancreatic acinar cell dedifferentiation avoiding cell death under stress conditions 1 2 2 1 1 1,3 Elyne Backx ● Elke Wauters ● Jonathan Baldan ● Mathias Van Bulck ● Ellis Michiels ● Yves Heremans ● 4 5 5 2 6 Diedert Luc De Paep ● Mineo Kurokawa ● Susumu Goyama ● Luc Bouwens ● Patrick Jacquemin ● 1,2 1 Isabelle Houbracken ● Ilse Rooman Received: 20 July 2020 / Revised: 3 March 2021 / Accepted: 4 March 2021 / Published online: 24 March 2021 © The Author(s) 2021. This article is published with open access Abstract Maintenance of the pancreatic acinar cell phenotype suppresses tumor formation. Hence, repetitive acute or chronic pancreatitis, stress conditions in which the acinar cells dedifferentiate, predispose for cancer formation in the pancreas. Dedifferentiated acinar cells acquire a large panel of duct cell-specific markers. However, it remains unclear to what extent dedifferentiated acini differ from native duct cells and which genes are uniquely regulating acinar cell dedifferentiation. Moreover, most studies have been performed on mice since the availability of human cells is scarce. Here, we applied a non- fi 1234567890();,: 1234567890();,: genetic lineage tracing method of human pancreatic exocrine acinar and duct cells that allowed cell-type-speci c gene expression profiling by RNA sequencing. Subsequent to this discovery analysis, one transcription factor that was unique for dedifferentiated acinar cells was functionally characterized. RNA sequencing analysis showed that human dedifferentiated acinar cells expressed genes in “Pathways of cancer” with a prominence of MECOM (EVI-1), a transcription factor that was not expressed by duct cells. -

Prospective Isolation of NKX2-1–Expressing Human Lung Progenitors Derived from Pluripotent Stem Cells
The Journal of Clinical Investigation RESEARCH ARTICLE Prospective isolation of NKX2-1–expressing human lung progenitors derived from pluripotent stem cells Finn Hawkins,1,2 Philipp Kramer,3 Anjali Jacob,1,2 Ian Driver,4 Dylan C. Thomas,1 Katherine B. McCauley,1,2 Nicholas Skvir,1 Ana M. Crane,3 Anita A. Kurmann,1,5 Anthony N. Hollenberg,5 Sinead Nguyen,1 Brandon G. Wong,6 Ahmad S. Khalil,6,7 Sarah X.L. Huang,3,8 Susan Guttentag,9 Jason R. Rock,4 John M. Shannon,10 Brian R. Davis,3 and Darrell N. Kotton1,2 2 1Center for Regenerative Medicine, and The Pulmonary Center and Department of Medicine, Boston University School of Medicine, Boston, Massachusetts, USA. 3Center for Stem Cell and Regenerative Medicine, Brown Foundation Institute of Molecular Medicine, University of Texas Health Science Center, Houston, Texas, USA. 4Department of Anatomy, UCSF, San Francisco, California, USA. 5Division of Endocrinology, Diabetes and Metabolism, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, Massachusetts, USA. 6Department of Biomedical Engineering and Biological Design Center, Boston University, Boston, Massachusetts, USA. 7Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, Massachusetts, USA. 8Columbia Center for Translational Immunology & Columbia Center for Human Development, Columbia University Medical Center, New York, New York, USA. 9Department of Pediatrics, Monroe Carell Jr. Children’s Hospital, Vanderbilt University, Nashville, Tennessee, USA. 10Division of Pulmonary Biology, Cincinnati Children’s Hospital, Cincinnati, Ohio, USA. It has been postulated that during human fetal development, all cells of the lung epithelium derive from embryonic, endodermal, NK2 homeobox 1–expressing (NKX2-1+) precursor cells. -

Factsheet XL T3;3 GATA2MECOM DF D-5124.Cdt
XL t(3;3) GATA2/ MECOM DF Translocation/Dual Fusion Order No.: Probe D-5124-100-OG Description XL t(3;3) GATA2/MECOM DF is designed as a dual fusion probe. The orange labeled probe spans the breakpoint at 3q26 (MECOM), the green labeled probe spans the breakpoint at 3q21 (GATA2 and RPN1). Clinical Details The chromosomal aberrations inv(3)(q21q26.2) and t(3;3)(q21;q26.2) characterize a distinct entity within patients with acute myeloid leukemia (AML). Their incidence in AML is about 1%-2.5% and patients have an unfavorable prognosis and low response to chemotherapy. Inv(3)/t(3;3) juxtapose the GATA2 enhancer with the MECOM locus which results in overexpression of EVI1. The EVI1 gene is located in 3q26 and is involved in hematopoietic stem cell maintenance. EVI1 is together with MDS located in the ´MDS1 and EVI1 complex locus´ (MECOM) and is the major player in this subtype XL t(3;3) GATA2/MECOM DF hybridized to lymphocytes. of AML. Furthermore, structural rearrangements caused by inv(3)/t(3;3) result in One normal interphase and two chromosomes 3 are reduced GATA2 expression which may contribute to the oncogenic potential of this shown. The expected normal signal pattern of XL t(3;3) GATA2/MECOM DF is two orange and two green aberration. GATA2 is located in chromosomal region 3q21 and is involved in develop- signals, representing the two normal MECOM and ment and proliferation of hematopoietic stem cells. Several other recurrent 3q26 G ATA 2 l o c i . B o t h , i n v ( 3 ) ( q 2 1 q 2 6 . -

Leukaemia Section
Atlas of Genetics and Cytogenetics in Oncology and Haematology INIST -CNRS OPEN ACCESS JOURNAL Leukaemia Section Short Communication t(3;12)(q26;p13) ETV6/MECOM / t(3;12)(q26;p13) ETV6/EVI1 Etienne De Braekeleer, Nathalie Douet-Guilbert, Marc De Braekeleer Cytogenetics Laboratory, Faculty of Medicine, University of Brest, France (EDB, NDG, MDB) Published in Atlas Database: April 2014 Online updated version : http://AtlasGeneticsOncology.org/Anomalies/t0312.html DOI: 10.4267/2042/54377 This article is an update of : Desangles F. t(3;12)(q26;p13). Atlas Genet Cytogenet Oncol Haematol 1997;1(1):21. This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence. © 2014 Atlas of Genetics and Cytogenetics in Oncology and Haematology Abstract M7. Short communication on t(3;12)(q26;p13) Epidemiology ETV6/MECOM, with data on clinics, and the genes 46 cases described so far; sex ratio: 29M/16F (1 implicated. unknown); age: 2.5-87 yrs (med: 49 yrs), unknown age for 10 cases. Identity Cytology Note Dysplasia of megakaryocytes, multilineage Only a few cases were shown to involve a involvement. ETV6/MECOM fusion by FISH or RT-PCR. In Prognosis other cases, only MECOM or ETV6 was shown to be rearranged by FISH. Over-expression of See survival curve above. MECOM/EVI1 was found in other cases. Finally, no cytogenetic or molecular analysis was performed Cytogenetics in a few cases. Cytogenetics molecular Heterogeneity of the EVI1 breakpoints, as evidenced by the Cytocell Aquarius EVI1 Breakapart probe. The EVI1 Breakapart probe contains three probes: a probe labeled in Aqua of 562 kb in size centromeric to the EVI1 gene, a probe labeled in Spectrum Green of 181 kb covering EVI1 and its flanking regions and a probe labeled in Spectrum Orange of 124 kb telomeric of the EVI1 gene (telomeric of t(3;12)(q26;p13) G-banding - Courtesy Jean-Luc Lai and Alain Vanderhaegen.Clinics and pathology MYNN and covering LRRC34). -

IGH Rearrangement in Myeloid Neoplasms Sion and Activation
CASE REPORTS genes such as MYC and BCL2, leading to their overexpres- IGH rearrangement in myeloid neoplasms sion and activation. Here, we found two IGH rearrange- ments in myeloid tumors, including an IGH-MECOM in a Though immunoglobulin genes are typically expressed myelodysplastic syndrome (MDS) and an IGH-CCNG1 in in B lymphocytes, recent studies found ectopic an AML. Our studies provide the first evidence that a once immunoglobulin expression in non-B-cell tumor cells believed B-cell tumor-specific oncogenic mechanism is including acute myeloid leukemia (AML). The also present in myeloid tumors. immunoglobulin genes, including immunoglobulin heavy Case #1: A 60-year-old female with a history of breast chain genes (IGH), light kappa (k) chain genes (IGK) and cancer presented with fatigue, bilateral flank pain, and 9 light lambda (λ) chain genes (IGL), are frequently hematuria. Complete blood count showed 24.82x10 /L of rearranged in B-cell tumors. These rearrangements result white blood cells, 11.2 g/dL of hemoglobin, 33.9% of in a juxtaposition of IG enhancers to the vicinity of onco- hematocrit, and 68x109/L of platelets. The bone marrow A B C D E Figure 1. Acute myeloid leukemia with an IGH-CCNG1 rearrangement. (A) Chromosome analysis of the bone marrow aspirate showed a complex karyotype, including an unbalanced translocation involving chromosomes 5, 14 and 19. Arrows indicate clonal aberrations. (B) Fluorescence in situ hybridization (FISH) on abnormal metaphase showed the 3'IGH (red) remaining on the derivative chromosome 14 and the 5'IGH lost, consistent with an unbalanced IGH rearrange- ment. -

Cryptic Genomic Lesions in Adverse-Risk Acute Myeloid Leukemia Identified by Integrated Whole Genome and Transcriptome Sequencing
Leukemia (2020) 34:306–311 https://doi.org/10.1038/s41375-019-0546-1 LETTER Cytogenetics and molecular genetics Cryptic genomic lesions in adverse-risk acute myeloid leukemia identified by integrated whole genome and transcriptome sequencing 1,2,3 2 3 3,4 2 Jaeseung C. Kim ● Philip C. Zuzarte ● Tracy Murphy ● Michelle Chan-Seng-Yue ● Andrew M. K. Brown ● 2 5 1,3,4 1,3 1,2,6 Paul M. Krzyzanowski ● Adam C. Smith ● Faiyaz Notta ● Mark D. Minden ● John D. McPherson Received: 15 March 2019 / Revised: 23 April 2019 / Accepted: 17 June 2019 / Published online: 21 August 2019 © The Author(s) 2019. This article is published with open access To the Editor: treated accordingly [1]. In recent years, advancements in next-generation sequencing and efforts by large genomics Acute myeloid leukemia (AML) is a heterogeneous group studies have led to a classification of 11 AML subgroups of hematologic malignancies characterized by the pro- based on cytogenetic abnormalities as well as mutations in liferation of myeloid cells blocked in their ability to dif- genes, such as NPM1 or CEBPA [1]. 1234567890();,: 1234567890();,: ferentiate. Evaluation by G-banding and fluorescence in situ AML with a complex karyotype, defined by the presence hybridization is an essential aspect in the initial disease of three or more unrelated chromosomal aberrations and the characterization and even now is fundamental in identifying absence of favorable cytogenetic rearrangements, is asso- cytogenetic abnormalities that can inform disease diagnosis, ciated with TP53 mutations and strikingly poor outcome prognosis, and treatment decision [1, 2]. For instance, the [1, 3]. -

4-Phenylthiobutanoic Acid Inhibits Histone Deacetylase 8 to Enhance Post-Acute Kidney Injury Repair
Title Page Slow and Steady: 4-Phenylthiobutanoic Acid Inhibits Histone Deacetylase 8 to Enhance Post-Acute Kidney Injury Repair by Hwa In Han B.S., College of William & Mary, 2014 Submitted to the Graduate Faculty of the School of Medicine in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Pittsburgh 2020 Committee Page UNIVERSITY OF PITTSBURGH SCHOOL OF MEDICINE This dissertation was presented by Hwa In Han It was defended on May 29, 2020 and approved by Jacqueline Ho, M.D., Department of Pediatric Nephrology Penelope Morel, M.D., Department of Immunology Kyle Orwig, Ph.D., Department of Obstetrics, Gynecology & Reproductive Services Michael Tsang, Ph.D., Department of Developmental Biology Dissertation Director: Neil Hukriede, Ph.D., Department of Developmental Biology ii Copyright © by Hwa In Han 2020 iii Abstract Slow and Steady: 4-Phenylthiobutanoic Acid Inhibits Histone Deacetylase 8 to Enhance Post-Acute Kidney Injury Repair Hwa In Han, PhD University of Pittsburgh, 2020 Acute kidney injury (AKI) is a rapid decline in kidney function that is associated with high mortality and morbidity rates, affecting 1 in 5 adults worldwide 1. While damaged renal tubular epithelial cells (RTECs) undergo reparative process to proliferate and replace the damaged epithelium, it is not a robust mechanism. The initial damage is followed by a cross-talk of innate immune system and RTECs resulting in exacerbation of inflammation, fibrosis, and sometimes irreversible damage. Despite the prevalence and severity of AKI, no FDA approved therapeutics exist to replace damaged or lost RTECS. Therefore, it is imperative to screen, identify, and understand mechanisms with which candidate therapeutics can enhance repair. -

Grimme, Acadia.Pdf
MECHANISM OF ACTION OF HISTONE DEACETYLASE INHIBITORS ON SURVIVAL MOTOR NEURON 2 PROMOTER by Acadia L. Grimme A thesis submitted to the Faculty of the University of Delaware in partial fulfillment of the requirements for the degree of Bachelors of Science in Biological Sciences with Distinction Spring 2018 © 2018 Acadia Grimme All Rights Reserved MECHANISM OF ACTION OF HISTONE DEACETYLASE INHIBITORS ON SURVIVAL MOTOR NEURON 2 PROMOTER by Acadia L. Grimme Approved: __________________________________________________________ Matthew E. R. Butchbach, Ph.D. Professor in charge of thesis on behalf of the Advisory Committee Approved: __________________________________________________________ Deni S. Galileo, Ph.D. Professor in charge of thesis on behalf of the Advisory Committee Approved: __________________________________________________________ Carlton R. Cooper, Ph.D. Committee member from the Department of Biological Sciences Approved: __________________________________________________________ Gary H. Laverty, Ph.D. Committee member from the Board of Senior Thesis Readers Approved: __________________________________________________________ Michael Chajes, Ph.D. Chair of the University Committee on Student and Faculty Honors ACKNOWLEDGMENTS I would like to acknowledge my thesis director Dr. Butchbach for his wonderful guidance and patience as I worked through my project. He has been an excellent research mentor over the last two years and I am forever thankful that he welcomed me into his lab. His dedication to his work inspires me as an aspiring research scientist. His lessons will carry on with me as I pursue future research in graduate school and beyond. I would like to thank both current and former members of the Motor Neuron Disease Laboratory: Sambee Kanda, Kyle Hinkle, and Andrew Connell. Sambee and Andrew patiently taught me many of the techniques I utilized in my project, and without them it would not be what it is today. -

Investigation of the Molecular Mechanisms Underlying Postoperative Recurrence in Prostate Cancer by Gene Expression Profiling
EXPERIMENTAL AND THERAPEUTIC MEDICINE 15: 761-768, 2018 Investigation of the molecular mechanisms underlying postoperative recurrence in prostate cancer by gene expression profiling CHENG YAJUN1*, TANG YUAN2*, WANG ZHONG1 and XU BIN1 1Department of Urology, Shanghai Ninth People's Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200011; 2Department of Gastrointestinal Surgery, Changzheng Hospital, Second Military Medical University, Shanghai 200003, P.R. China Received March 16, 2017; Accepted October 20, 2017 DOI: 10.3892/etm.2017.5510 Abstract. The present study aimed to identify potential genes The results of the present study have provided novel ideas for associated with prostate cancer (PCa) recurrence following predicting the prognosis of patients with PCa following RP. radical prostatectomy (RP) in order to improve the prediction of the prognosis of patients with PCa. The GSE25136 micro- Introduction array dataset, including 39 recurrent and 40 non-recurrent PCa samples, was downloaded from the Gene Expression Omnibus Prostate cancer (PCa), as one of the most common men's malig- database. Differentially-expressed genes (DEGs) were identi- nancy in America, is the second leading cause of cancer-related fied using limma packages, and the pheatmap package was death in men (1). Although more than 80% of PCa was diag- used to present the DEGs screened using a hierarchical cluster nosed as localized disease and commonly treated by radical analysis. Furthermore, gene ontology functional enrich- prostatectomy (RP), postoperative recurrence occurred in ment analysis was used to predict the potential functions of about 15% of patients within 5 years and up to 40% within the DEGs. Subsequently, Kyoto Encyclopedia of Genes and 10 years (2). -
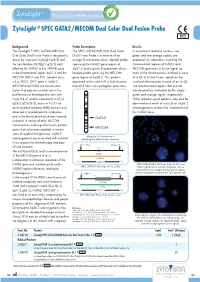
Zytolight ® SPEC GATA2/MECOM Dual Color Dual Fusion Probe
ZytoLight ® SPEC GATA2/MECOM Dual Color Dual Fusion Probe Background Probe Description Results The ZytoLight ® SPEC GATA2/MECOM The SPEC GATA2/MECOM Dual Color In a normal interphase nucleus, two Dual Color Dual Fusion Probe is designed to Dual Fusion Probe is a mixture of an green and two orange signals are detect the inversion inv(3)(q21q26.2) and orange fluorochrome direct labeled probe expected. An aberration involving the the translocation t(3;3)(q21;q26.2) both spanning the GATA2 gene region at chromosomal regions of GATA2 and affecting the GATA2 (a.k.a. NFE1B) gene 3q21.3 and a green fluorochrome direct MECOM generates a fusion signal on in the chromosomal region 3q21.3 and the labeled probe spanning the MECOM each of the chromosomes involved in case MECOM (MDS1 and EVI1 complex locus, gene region at 3q26.2. This probe is of a t(3;3) or two fusion signals on the a.k.a. MDS1, EVI1) gene in 3q26.2. approved to be used with a hybridization involved chromosome in case of an inv(3). MECOM and GATA2 are transcription time of 2 hours on cytological specimens. The chromosomal regions that are not factors that play an essential role in the translocated are indicated by the single proliferation of hematopoietic stem cells. green and orange signal, respectively. Inv(3)/t(3;3), and less commonly ins(3;3) Other relevant signal patterns may also be (q26.2;q21q26.2), occur in 1-2.5% of observed as a result of ins(3;3) or 3q26.2 acute myeloid leukemia (AML) and are also rearrangements without the involvement of observed in myelodysplastic syndromes the GATA2 locus. -

Hemavision®-28N Screening Test for 28 Chromosome Translocations and More Than 145 Breakpoints Associated with Leukemia
User manual HemaVision®-28N Screening test for 28 chromosome translocations and more than 145 breakpoints associated with leukemia USER MANUAL Cat No. HV01-28N DNA Diagnostic A/S www.dna-diagnostic.com Revision 2019.01.10 IVD HemaVisionÒ-28N Multiplex RT-PCR test Screens for 28 leukemia causing translocations del1(p32) (STIL-TAL1) t(9;12) (q34;p13) (ETV6-ABL1) t(1;11) (p32;q23.3) (KMT2A-EPS15) t(9,22) (q34;q11) (BCR-ABL1) t(1;11) (q21;q23.3) (KMT2A-MLLT11) t(10;11) (p12;q23.3) (KMT2A-MLLT10) t(1;19) (q23;p13) (TCF3-PBX1) t(11;17) (q23.3;q21) (KMT2A-MLLT6) t(3;5) (q25;q34) (NPM1-MLF1) t(11;17) (q23;q21) (ZBTB16-RARA) t(3;21) (q26;q22) (RUNX1-MECOM) t(11;19) (q23.3;p13.1) (KMT2A-ELL) t(4;11) (q21;q23.3) (KMT2A-AFF1) t(11;19) (q23.3;p13.3) (KMT2A-MLLT1) t(5;12) (q33;p13) (ETV6-PDGFRB) t(12;21) (p13;q22) (ETV6-RUNX1) t(5;17) (q35;q21) (NPM1-RARA) t(12;22) (p13;q11) (ETV6-MN1) t(6;9) (p23;q34) (DEK-NUP214) t(15;17) (q24;q21) (PML-RARA) t(6;11) (q27;q23.3) (KMT2A-AFDN) inv(16) (p13;q22) (CBFB-MYH11) t(8;21) (q22;q22) (RUNX1-RUNX1T1) t(16;21) (p11;q22) (FUS-ERG) t(9;9) (q34;q34) (SET-NUP214) t(17;19) (q22;p13) (TCF3-HLF) t(9;11) (p21.3;q23.3) (KMT2A-MLLT3) t(X;11) (q13;q23.3) (KMT2A-FOXO4) USER MANUAL for HemaVisionÒ-28N Cat. -

Leukaemia Section
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL INIST-CNRS Leukaemia Section Short Communication inv(3)(q21q26) RPN1/MECOM / t(3;3)(q21;q26) RPN1/MECOM / ins(3;3)(q26;q21q26) RPN1/MECOM Thomas Smo Laboratoire d'Hematologie-Immunologie-Cytogenetique, Centre Hospitalier de Valenciennes, France Published in Atlas Database: November 2014 Online updated version : http://AtlasGeneticsOncology.org/Anomalies/inv3ID1006.html Printable original version : http://documents.irevues.inist.fr/bitstream/handle/2042/62326/11-2014-inv3ID1006.pdf DOI: 10.4267/2042/62326 This article is an update of : inv(3)(q21q26) RPN1/MECOM. Atlas Genet Cytogenet Oncol Haematol 2015;19(9) Huret JL. inv(3)(q21q26). Atlas Genet Cytogenet Oncol Haematol 1997;1(2) This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence. © 2015 Atlas of Genetics and Cytogenetics in Oncology and Haematology common 3q abnormalities in AML (32%). Abstract The frequency of these rearrangements is estimated Review on inv(3)(q21q26) RPN1/MECOM, with to range between 1.4% and 1.6% of AML in adults data on clinics, and the genes implicated. with no difference between sexes. These rearrangements are slightly more common in Identity patients aged 60 years or younger, and extremely rare in pediatric AML. Note The three chromosome anomalies are variants of Clinics each other, and they share identical features. Patients may present a normal platelet count, however marked thrombocytosis may occur in 7% to Clinics and pathology 22% of patients. Disease Cytology Blasts express CD13, CD33, CD117, HLA-DR, inv(3) and t(3;3) have been documented in de novo CD56, CD34 and CD38; CD7 is aberrantly AML (in all FAB subtypes except M3), t-AML, s- expressed in some cases, whereas the other lymphoid AML, myelodysplastic syndrome (MDS), chronic markers are uncommon; blasts may also express myelogenous leukaemia (CML), more often in megakaryocytic markers such as CD41 or CD61.