Crystallographic and Cryogenic Electron Microscopic Structures And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Metals, Minerals and Microbes: Geomicrobiology and Bioremediation
Microbiology (2010), 156, 609–643 DOI 10.1099/mic.0.037143-0 SGM Prize Metals, minerals and microbes: geomicrobiology Lecture and bioremediation Geoffrey Michael Gadd Division of Molecular Microbiology, College of Life Sciences, University of Dundee, Dundee Correspondence DD1 5EH, UK Geoffrey Michael Gadd [email protected] Microbes play key geoactive roles in the biosphere, particularly in the areas of element biotransformations and biogeochemical cycling, metal and mineral transformations, decomposition, bioweathering, and soil and sediment formation. All kinds of microbes, including prokaryotes and eukaryotes and their symbiotic associations with each other and ‘higher organisms’, can contribute actively to geological phenomena, and central to many such geomicrobial processes are transformations of metals and minerals. Microbes have a variety of properties that can effect changes in metal speciation, toxicity and mobility, as well as mineral formation or mineral dissolution or deterioration. Such mechanisms are important components of natural biogeochemical cycles for metals as well as associated elements in biomass, soil, rocks and minerals, e.g. sulfur and phosphorus, and metalloids, actinides and metal radionuclides. Apart from being important in natural biosphere processes, metal and mineral transformations can have beneficial or detrimental consequences in a human context. Bioremediation is the application of biological systems to the clean-up of organic and inorganic pollution, with bacteria and fungi being the most important organisms for reclamation, immobilization or detoxification of metallic and radionuclide pollutants. Some biominerals or metallic elements deposited by microbes have catalytic and other properties in nanoparticle, crystalline or colloidal forms, and these are relevant to the development of novel biomaterials for technological and antimicrobial purposes. -

4 Metabolic and Taxonomic Diversification in Continental Magmatic Hydrothermal Systems
Maximiliano J. Amenabar, Matthew R. Urschel, and Eric S. Boyd 4 Metabolic and taxonomic diversification in continental magmatic hydrothermal systems 4.1 Introduction Hydrothermal systems integrate geological processes from the deep crust to the Earth’s surface yielding an extensive array of spring types with an extraordinary diversity of geochemical compositions. Such geochemical diversity selects for unique metabolic properties expressed through novel enzymes and functional characteristics that are tailored to the specific conditions of their local environment. This dynamic interaction between geochemical variation and biology has played out over evolu- tionary time to engender tightly coupled and efficient biogeochemical cycles. The timescales by which these evolutionary events took place, however, are typically in- accessible for direct observation. This inaccessibility impedes experimentation aimed at understanding the causative principles of linked biological and geological change unless alternative approaches are used. A successful approach that is commonly used in geological studies involves comparative analysis of spatial variations to test ideas about temporal changes that occur over inaccessible (i.e. geological) timescales. The same approach can be used to examine the links between biology and environment with the aim of reconstructing the sequence of evolutionary events that resulted in the diversity of organisms that inhabit modern day hydrothermal environments and the mechanisms by which this sequence of events occurred. By combining molecu- lar biological and geochemical analyses with robust phylogenetic frameworks using approaches commonly referred to as phylogenetic ecology [1, 2], it is now possible to take advantage of variation within the present – the distribution of biodiversity and metabolic strategies across geochemical gradients – to recognize the extent of diversity and the reasons that it exists. -

The Euryarchaeotes, a Subdomain of Archaea, Survive on a Single DNA Polymerase: Fact Or Farce?
Genes Genet. Syst. (1998) 73, p. 323–336 REVIEW The euryarchaeotes, a subdomain of Archaea, survive on a single DNA polymerase: Fact or farce? Yoshizumi Ishino* and Isaac K. O. Cann Department of Molecular Biology, Biomolecular Engineering Research Institute, 6-2-3 Furuedai, Suita, Osaka 565-0874, Japan Archaea is now recognized as the third domain of life. Since their discovery, much effort has been directed towards understanding the molecular biology and biochemistry of Archaea. The objective is to comprehend the complete structure and the depth of the phylogenetic tree of life. DNA replication is one of the most important events in living organisms and DNA polymerase is the key enzyme in the molecular machinery which drives the process. All archaeal DNA polymerases were thought to belong to family B. This was because all of the products of pol genes that had been cloned showed amino acid sequence similarities to those of this family, which includes three eukaryal DNA replicases and Escherichia coli DNA polymerase II. Recently, we found a new heterodimeric DNA polymerase from the hyperthermophilic archaeon, Pyrococcus furiosus. The genes coding for the subunits of this DNA polymerase are conserved in the euryarchaeotes whose genomes have been completely sequenced. The biochemical characteristics of the novel DNA polymerase family suggest that its members play an important role in DNA replication within euryarchaeal cells. We review here our current knowledge on DNA polymerases in Archaea with emphasis on the novel DNA polymerase discovered in Euryarchaeota. arily distinct from the Bacteria and rather appear to INTRODUCTION share a common ancestor with the Eukarya. -
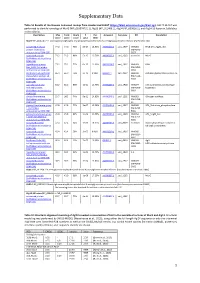
Supplementary Data
Supplementary Data Table S1 Results of the Domain Enhanced Lookup Time Accelerated BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). DELTA BLAST was performed to identify homologs of MurG (WP_063074721.1), Alg13 (NP_011468.1), Alg14 (NP_009626.1), and Alg14-13 fusion in Sulfolobus acidocaldarius. Description Max Total Query E Per. Accession Saci gene KO Annotation score score cover value Ident MurG WP_063074721.1 undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase [Escherichia coli] conserved Archaeal 77.0 77.0 76% 6e-17 14.52% AAY81210.1 saci_1907 MW061 RFaB GT1_YqgM_like protein [Sulfolobus BM-A045- acidocaldarius DSM 639] B027 conserved protein 71.2 71.2 98% 5e-15 16.76% AAY80607.1 saci_1262 essential MurG [Sulfolobus acidocaldarius DSM 639] hypothetical protein 71.2 71.2 72% 6e-15 13.33% AAY81224.1 saci_1921 MW069 RfaB Saci_1921 [Sulfolobus BM-A055- acidocaldarius DSM 639] B003 RecName: Full=Archaeal 65.9 65.9 75% 3e-13 9.90% Q4JAK2.1 saci_0807 MW043 Archaeal glycosylation protein 16 glycosylation protein 16 BM-A110- [Sulfolobus acidocaldarius B008 DSM 639] partially conserved 61.2 61.2 96% 1e-11 12.76% AAY81207.1 saci_1904 MW077 Cell wall/membrane/envelope Archaeal protein BM-A050- biogenesis [Sulfolobus acidocaldarius B002 DSM 639] glycosyl transferase 59.7 98.2 74% 5e-11 14.02% AAY80548.1 saci_1201 MW053 Glycogen synthase [Sulfolobus acidocaldarius BM-A150- DSM 639] B1 glycosyl transferase group 47.8 47.8 77% 3e-07 12.38% AAY81133.1 saci_1827 MW042 GT1_Trehalose_phosphorylase 1 [Sulfolobus BM-A280- acidocaldarius -

Virus-Borne Mini-CRISPR Arrays Are Involved in Interviral Conflicts
ARTICLE https://doi.org/10.1038/s41467-019-13205-2 OPEN Virus-borne mini-CRISPR arrays are involved in interviral conflicts Sofia Medvedeva1,2,3, Ying Liu1, Eugene V. Koonin 4, Konstantin Severinov2,5,6, David Prangishvili1,7 & Mart Krupovic 1* CRISPR-Cas immunity is at the forefront of antivirus defense in bacteria and archaea and specifically targets viruses carrying protospacers matching the spacers catalogued in the 1234567890():,; CRISPR arrays. Here, we perform deep sequencing of the CRISPRome—all spacers contained in a microbiome—associated with hyperthermophilic archaea of the order Sulfolobales recovered directly from an environmental sample and from enrichment cultures established in the laboratory. The 25 million CRISPR spacers sequenced from a single sampling site dwarf the diversity of spacers from all available Sulfolobales isolates and display complex temporal dynamics. Comparison of closely related virus strains shows that CRISPR targeting drives virus genome evolution. Furthermore, we show that some archaeal viruses carry mini-CRISPR arrays with 1–2 spacers and preceded by leader sequences but devoid of cas genes. Closely related viruses present in the same population carry spacers against each other. Targeting by these virus-borne spacers represents a distinct mechanism of heterotypic superinfection exclusion and appears to promote archaeal virus speciation. 1 Institut Pasteur, Department of Microbiology, 75015 Paris, France. 2 Center of Life Sciences, Skolkovo Institute of Science and Technology, Skolkovo, Russia. 3 Sorbonne Université, Collège doctoral, 75005 Paris, France. 4 National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD 20894, USA. 5 Waksman Institute, Rutgers University, Piscataway, NJ 08854, USA. 6 Institute of Molecular Genetics, Moscow 123182, Russia. -

Downloaded (July 2018) and Aligned Using Msaprobs V0.9.7 (16)
bioRxiv preprint doi: https://doi.org/10.1101/524215; this version posted January 20, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Positively twisted: The complex evolutionary history of Reverse Gyrase suggests a non- hyperthermophilic Last Universal Common Ancestor Ryan Catchpole1,2 and Patrick Forterre1,2 1Institut Pasteur, Unité de Biologie Moléculaire du Gène chez les Extrêmophiles (BMGE), Département de Microbiologie F-75015 Paris, France 2Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, Univ. Paris-Sud, Univ. Paris-Saclay, 91198, Gif-sur-Yvette Cedex, France 1 bioRxiv preprint doi: https://doi.org/10.1101/524215; this version posted January 20, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Reverse gyrase (RG) is the only protein found ubiquitously in hyperthermophilic organisms, but absent from mesophiles. As such, its simple presence or absence allows us to deduce information about the optimal growth temperature of long-extinct organisms, even as far as the last universal common ancestor of extant life (LUCA). The growth environment and gene content of the LUCA has long been a source of debate in which RG often features. In an attempt to settle this debate, we carried out an exhaustive search for RG proteins, generating the largest RG dataset to date. -

Resolving Widespread Incomplete and Uneven Archaeal Classifications
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.01.972265; this version posted February 17, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 2 Resolving widespread incomplete and uneven archaeal 3 classifications based on a rank-normalized genome-based 4 taxonomy 5 Christian Rinke1, Maria Chuvochina1, Aaron J. Mussig1, Pierre-Alain Chaumeil1, Adrian A. Davin1, David W. 6 Waite2, William B Whitman3, Donovan H. Parks1, Philip Hugenholtz1 7 8 1 Australian Centre for Ecogenomics, School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, Australia 9 2 School of Biological Sciences, The University of Auckland, New Zealand 10 3 Department of Microbiology, University of Georgia Athens, GA, USA 11 12 Abstract 13 An increasing wealth of genomic data from cultured and uncultured microorganisms provides the opportunity 14 to develop a systematic taxonomy based on evolutionary relationships. Here we propose a standardized 15 archaeal taxonomy, as part of the Genome Taxonomy Database (GTDB), derived from a 122 concatenated 16 protein phylogeny that resolves polyphyletic groups and normalizes ranks based on relative evolutionary 17 divergence (RED). The resulting archaeal taxonomy is stable under a range of phylogenetic variables, 18 including marker genes, inference methods, corrections for rate heterogeneity and compositional bias, tree 19 rooting scenarios, and expansion of the genome database. Rank normalization was shown to robustly correct 20 for substitution rates varying up to 30-fold using simulated datasets. -

Application for Approval to Import Into Containment Any New Organism That
ER-AN-02N 10/02 Application for approval to import into FORM 2N containment any new organism that is not genetically modified, under Section 40 of the Page 1 Hazardous Substances and New Organisms Act 1996 FORM NO2N Application for approval to IMPORT INTO CONTAINMENT ANY NEW ORGANISM THAT IS NOT GENETICALLY MODIFIED under section 40 of the Hazardous Substances and New Organisms Act 1996 Application Title: Importation of extremophilic microorganisms from geothermal sites for research purposes Applicant Organisation: Institute of Geological & Nuclear Sciences ERMA Office use only Application Code: Formally received:____/____/____ ERMA NZ Contact: Initial Fee Paid: $ Application Status: ER-AN-02N 10/02 Application for approval to import into FORM 2N containment any new organism that is not genetically modified, under Section 40 of the Page 2 Hazardous Substances and New Organisms Act 1996 IMPORTANT 1. An associated User Guide is available for this form. You should read the User Guide before completing this form. If you need further guidance in completing this form please contact ERMA New Zealand. 2. This application form covers importation into containment of any new organism that is not genetically modified, under section 40 of the Act. 3. If you are making an application to import into containment a genetically modified organism you should complete Form NO2G, instead of this form (Form NO2N). 4. This form, together with form NO2G, replaces all previous versions of Form 2. Older versions should not now be used. You should periodically check with ERMA New Zealand or on the ERMA New Zealand web site for new versions of this form. -

Supplementary Material For: Undinarchaeota Illuminate The
Supplementary Material for: Undinarchaeota illuminate the evolution of DPANN archaea Nina Dombrowski1, Tom A. Williams2, Benjamin J. Woodcroft3, Jiarui Sun3, Jun-Hoe Lee4, Bui Quang MinH5, CHristian Rinke5, Anja Spang1,5,# 1NIOZ, Royal NetHerlands Institute for Sea ResearcH, Department of Marine Microbiology and BiogeocHemistry, and UtrecHt University, P.O. Box 59, NL-1790 AB Den Burg, THe NetHerlands 2 ScHool of Biological Sciences, University of Bristol, Bristol, BS8 1TQ, UK 3Australian Centre for Ecogenomics, ScHool of CHemistry and Molecular Biosciences, THe University of Queensland, QLD 4072, Australia 4Department of Cell- and Molecular Biology, Science for Life Laboratory, Uppsala University, SE-75123, Uppsala, Sweden 5ResearcH ScHool of Computer Science and ResearcH ScHool of Biology, Australian National University, ACT 2601, Australia #corresponding autHor. Postal address: Landsdiep 4, 1797 SZ 't Horntje (Texel). Email address: [email protected]. PHone number: +31 (0)222 369 526 Table of Contents Table of Contents 2 General 3 Evaluating CHeckM completeness estimates 3 Screening for contaminants 3 Phylogenetic analyses 4 Informational processing and repair systems 7 Replication and cell division 7 Transcription 7 Translation 8 DNA-repair and modification 9 Stress tolerance 9 Metabolic features 10 Central carbon and energy metabolism 10 Anabolism 13 Purine and pyrimidine biosyntHesis 13 Amino acid degradation and biosyntHesis 14 Lipid biosyntHesis 15 Vitamin and cofactor biosyntHesis 16 Host-symbiont interactions 16 Genes potentially -

Physiological Characterization of Sulfolobus Acidocaldarius in a Controlled Bioreactor Environment
International Journal of Environmental Research and Public Health Article Physiological Characterization of Sulfolobus acidocaldarius in a Controlled Bioreactor Environment Kerstin Rastädter , David Johannes Wurm , Oliver Spadiut and Julian Quehenberger * Research Division Biochemical Engineering, Faculty of Technical Chemistry, Institute of Chemical, Environmental and Bioscience Engineering, TU Wien, 1060 Vienna, Austria; [email protected] (K.R.); [email protected] (D.J.W.); [email protected] (O.S.) * Correspondence: [email protected] Abstract: The crenarchaeal model organism Sulfolobus acidocaldarius is typically cultivated in shake flasks. Although shake flasks represent the state-of-the-art for the cultivation of this microorganism, in these systems crucial process parameters, like pH or substrate availability, are only set initially, but cannot be controlled during the cultivation process. As a result, a thorough characterization of growth parameters under controlled conditions is still missing for S. acidocaldarius. In this study, we conducted chemostat cultivations at 75 ◦C using a growth medium containing L-glutamate and D-glucose as main carbon sources. Different pH values and dilution rates were applied with the goal to physiologically characterize the organism in a controlled bioreactor environment. Under these controlled conditions a pH optimum of 3.0 was determined. Washout of the cells occurred at a dilution rate of 0.097 h−1 and the optimal productivity of biomass was observed at a dilution rate −1 of 0.062 h . While both carbon sources were taken up by S. acidocaldarius concomitantly, a 6.6-fold higher affinity for L-glutamate was shown. When exposed to suboptimal growth conditions, S. -
Enzymatic Antioxidant Signatures in Hyperthermophilic Archaea
antioxidants Review Enzymatic Antioxidant Signatures in Hyperthermophilic Archaea Emilia Pedone 1, Gabriella Fiorentino 2 , Simonetta Bartolucci 2 and Danila Limauro 2,* 1 Istituto di Biostrutture e Bioimmagini, CNR, Via Mezzocannone 16, 80134 Napoli, Italy; [email protected] 2 Dipartimento di Biologia, Università degli Studi di Napoli Federico II, Complesso universitario Monte S. Angelo, Via Cinthia, 80126 Napoli, Italy; fi[email protected] (G.F.); [email protected] (S.B.) * Correspondence: [email protected] Received: 6 July 2020; Accepted: 31 July 2020; Published: 3 August 2020 Abstract: To fight reactive oxygen species (ROS) produced by both the metabolism and strongly oxidative habitats, hyperthermophilic archaea are equipped with an array of antioxidant enzymes whose role is to protect the biological macromolecules from oxidative damage. The most common ROS, such as superoxide radical (O2•−) and hydrogen peroxide (H2O2), are scavenged by superoxide dismutase, peroxiredoxins, and catalase. These enzymes, together with thioredoxin, protein disulfide oxidoreductase, and thioredoxin reductase, which are involved in redox homeostasis, represent the core of the antioxidant system. In this review, we offer a panorama of progression of knowledge on the antioxidative system in aerobic or microaerobic (hyper)thermophilic archaea and possible industrial applications of these enzymes. Keywords: antioxidant enzymes; oxidative stress; reactive oxygen species; hyperthermophiles; archaea 1. Introduction The first forms of life evolved in an anaerobic and reducing environment, in which the main gases in the atmosphere were represented by H2, CH4, and NH3. Due to the photosynthetic activity of cyanobacteria, the accumulation of oxygen imposed a great selective pressure favoring the development of more complex life forms c.a. -
Flexibility in the Mineral Dependent Metabolism of A
FLEXIBILITY IN THE MINERAL DEPENDENT METABOLISM OF A THERMOACIDOPHILIC CRENARCHAEOTE by Maximiliano Jose Amenabar Barriuso A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Microbiology MONTANA STATE UNIVERSITY Bozeman, Montana August 2017 ©COPYRIGHT by Maximiliano Jose Amenabar Barriuso 2017 All Rights Reserved ii DEDICATION For my wife Ivonne who has been the pillar of support and love in my life and for my daughters Martina and Trinidad who are the ones that inspire my life every day. iii ACKNOWLEDGEMENTS I would like to extend my gratitude to Dr. Eric Boyd for all his time and patience in guiding me through all my Ph.D. It is hard to express my gratitude to him in just a few words, but I just could say that I have been really lucky to learn from him. I also would like to express my thanks to Dr. John Peters for giving me the opportunity to join his lab and making my life easier in applying and attending Montana State University. I am also thankful to everyone in the Boyd lab: Dan, John, Melody, Saroj, Eric D., Erik A., Libby and former lab member Matt Urschel for being such a fun and smart group of people to work with. Thanks also to the various co-authors who contributed to the chapters of these dissertation and to my committee members: Dr. Matthew Fields, Dr. Mark Skidmore and Dr. John Peters; all of you helped me grow as a scientist. I am also thankful to the Department of Microbiology and Immunology for believing in me and giving me the opportunity to join the Department to do my PhD, and also to all the former and current staff from the Department for helping me any time I need it.