Optochin Resistance in Streptococcus Pneumoniae: Mechanism, Significance, and Clinical Implications
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Prevalence of Urinary Tract Infection and Antibiotic Resistance Pattern in Pregnant Women, Najran Region, Saudi Arabia
Vol. 13(26), pp. 407-413, August, 2019 DOI: 10.5897/AJMR2019.9084 Article Number: E3F64FA61643 ISSN: 1996-0808 Copyright ©2019 Author(s) retain the copyright of this article African Journal of Microbiology Research http://www.academicjournals.org/AJMR Full Length Research Paper Prevalence of urinary tract infection and antibiotic resistance pattern in pregnant women, Najran region, Saudi Arabia Ali Mohamed Alshabi1*, Majed Saeed Alshahrani2, Saad Ahmed Alkahtani1 and Mohammad Shabib Akhtar1 1Department of Clinical Pharmacy, College of Pharmacy, Najran University, Najran, Saudi Arabia. 2Department of Obstetics and Gyneocology, Faculty of Medicine, Najran University, Najran, Saudi Arabia. Received 25 February, 2019; Accepted August 5, 2019 Urinary Tract Infection (UTI) is one of the commonest infectious disease in pregnancy, and in pregnancy we have very limited number of antibiotics to treat the UTI. This study was conducted on 151 patients who attended the gynecology clinic during the study period. Nineteen UTI proven cases of UTI were studied for prevalence of microorganism and sensitivity pattern against different antibiotics. Among the bacteria isolated, Escherichia coli (73.68%) and Staphylococcus aureus (10.52%) were the most prevalent Gram negative and Gram positive bacteria respectively. To know the resistance pattern of microorganism we used commercially available discs of different antibiotics. Gram negative bacteria showed more resistance as compared to Gram positive one. It is observed that the most effective antibiotic for Gram negative isolates is Ceftriaxone (87.5%), followed by Amoxicillin + Clavulanic acid (81.25%), Amikacin (75%), Cefuroxime (75%), Cefixime (68.75%) and Mezlocillin (62.5%). For the Gram positive bacteria, Ceftriaxone, Amikacin and Amoxicillin + Clavulanic acid were the most effective antimicrobials (100%). -

Antimicrobial Activity and Antibiotic Resistance of Lactobacillus Delbrueckii Ssp
African Journal of Microbiology Research Vol. 5(6) pp. 675-682, 18 March, 2011 Available online http://www.academicjournals.org/ajmr DOI: 10.5897/AJMR10.835 ISSN 1996-0808 ©2011 Academic Journals Full Length Research Paper Antimicrobial activity and antibiotic resistance of Lactobacillus delbrueckii ssp. bulgaricus and Streptococcus thermophilus strains isolated from Turkish homemade yoghurts Asli Akpinar, Oktay Yerlikaya* and Sevda Kiliç Department of Dairy Technology, Faculty of Agriculture, Ege University, 35100, Bornova – zmir, Turkey. Accepted 1 March, 2011 The aim of this study was to determine the inhibitive effect of 25 Lactobacillus delbruecki ssp. bulgaricus and 16 Streptococcus thermophilus strains isolated from 30 different homemade yoghurts on several pathogen and contaminant bacteria. The antibiotic resistance of these bacteria was also determined. All of Lactobacillus bulgaricus strains exhibited antimicrobial activity against Escherichia coli, whereas all of S. thermophilus strains exhibited the same activity against Klebsiella pneumoniae. None of L. bulgaricus strains were resistant to the polymixin-B, only the OL4 strain has shown resistance to bacitracin. While some strains of S. thermophilus like C6 and SL4 exhibited resistance to novobiocin, SY72, M3, C1M, and F1M were shown to optochin. ET6 and SY73 strains were found to be resistant in both novobiocin and optochin. Key words: Antibiotic resistance, antimicrobial activity, homemade yoghurt, lactic acid bacteria, Lactobacillus delbrueckii ssp. bulgaricus, Streptococcus thermophilus. INTRODUCTION Generally, yoghurt is being perceived as a kind of healthy properties and they have a long history of safe use as food which has a low fat and essential value in terms of starter culture bacteria (Katla et al., 2001). -

Use of the Diagnostic Bacteriology Laboratory: a Practical Review for the Clinician
148 Postgrad Med J 2001;77:148–156 REVIEWS Postgrad Med J: first published as 10.1136/pmj.77.905.148 on 1 March 2001. Downloaded from Use of the diagnostic bacteriology laboratory: a practical review for the clinician W J Steinbach, A K Shetty Lucile Salter Packard Children’s Hospital at EVective utilisation and understanding of the Stanford, Stanford Box 1: Gram stain technique University School of clinical bacteriology laboratory can greatly aid Medicine, 725 Welch in the diagnosis of infectious diseases. Al- (1) Air dry specimen and fix with Road, Palo Alto, though described more than a century ago, the methanol or heat. California, USA 94304, Gram stain remains the most frequently used (2) Add crystal violet stain. USA rapid diagnostic test, and in conjunction with W J Steinbach various biochemical tests is the cornerstone of (3) Rinse with water to wash unbound A K Shetty the clinical laboratory. First described by Dan- dye, add mordant (for example, iodine: 12 potassium iodide). Correspondence to: ish pathologist Christian Gram in 1884 and Dr Steinbach later slightly modified, the Gram stain easily (4) After waiting 30–60 seconds, rinse with [email protected] divides bacteria into two groups, Gram positive water. Submitted 27 March 2000 and Gram negative, on the basis of their cell (5) Add decolorising solvent (ethanol or Accepted 5 June 2000 wall and cell membrane permeability to acetone) to remove unbound dye. Growth on artificial medium Obligate intracellular (6) Counterstain with safranin. Chlamydia Legionella Gram positive bacteria stain blue Coxiella Ehrlichia Rickettsia (retained crystal violet). -

Medical Bacteriology
LECTURE NOTES Degree and Diploma Programs For Environmental Health Students Medical Bacteriology Abilo Tadesse, Meseret Alem University of Gondar In collaboration with the Ethiopia Public Health Training Initiative, The Carter Center, the Ethiopia Ministry of Health, and the Ethiopia Ministry of Education September 2006 Funded under USAID Cooperative Agreement No. 663-A-00-00-0358-00. Produced in collaboration with the Ethiopia Public Health Training Initiative, The Carter Center, the Ethiopia Ministry of Health, and the Ethiopia Ministry of Education. Important Guidelines for Printing and Photocopying Limited permission is granted free of charge to print or photocopy all pages of this publication for educational, not-for-profit use by health care workers, students or faculty. All copies must retain all author credits and copyright notices included in the original document. Under no circumstances is it permissible to sell or distribute on a commercial basis, or to claim authorship of, copies of material reproduced from this publication. ©2006 by Abilo Tadesse, Meseret Alem All rights reserved. Except as expressly provided above, no part of this publication may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photocopying, recording, or by any information storage and retrieval system, without written permission of the author or authors. This material is intended for educational use only by practicing health care workers or students and faculty in a health care field. PREFACE Text book on Medical Bacteriology for Medical Laboratory Technology students are not available as need, so this lecture note will alleviate the acute shortage of text books and reference materials on medical bacteriology. -

MALDI‑TOF MS‑Based Direct‑On‑Target Microdroplet Growth Assay: Latest Developments
EXPERIMENTAL AND THERAPEUTIC MEDICINE 20: 2555-2556, 2020 COMMENT MALDI‑TOF MS‑based direct‑on‑target microdroplet growth assay: Latest developments IOANNIS K. NEONAKIS1 and DEMETRIOS A. SPANDIDOS2 1Department of Clinical Microbiology and Microbial Pathogenesis, University Hospital of Heraklion, 71201 Heraklion; 2Laboratory of Clinical Virology, School of Medicine, University of Crete, 71003 Heraklion, Greece Received June 19, 2020; Accepted July 8, 2020 DOI: 10.3892/etm.2020.8976 Abstract. The matrix-assisted laser desorption-ionization in each droplet spot is known, the MIC can be evaluated and time‑of‑flight mass spectrometry direct‑on‑target microdro- the microorganism can be characterized as susceptible or not. plet growth assay for the rapid susceptibility testing and the With this assay and using a panel of different antibiotics, not detection of the underlying antibiotic resistance mechanisms only can the susceptibility status of an isolate be determined of microbia has been recently introduced. In the present study, but also the underlining resistance mechanisms be revealed. we review the latest developments in the field. In the initial reports, the assay was used for the detection of Recently, the matrix‑assisted laser desorption‑ionization carbapenem nonsusceptibility in Gram‑negative pathogens time‑of‑flight mass spectrometry (MALDI‑TOF MS) both from colonies (1) and directly from positive blood cultures direct‑on‑target microdroplet growth assay (DOT‑MGA) for (BCs) (2). The assay was also used for the detection of two the rapid susceptibility testing and the detection of the under- additional resistance mechanisms among Enterobacterales, lying antibiotic resistance mechanisms of microbia has been namely the extended‑spectrum β‑lactamase (ESBL) produc- introduced (1‑3). -
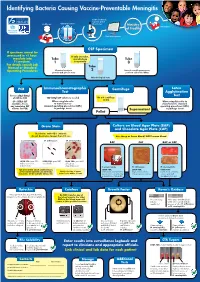
Identifying Bacteria Causing Vaccine-Preventable Meningitis
Identifying Bacteria Causing Vaccine-Preventable Meningitis Sentinel, national, regional, global Health care Clinical specimen laboratories Ministry Vaccination of Health Data management CSF Specimen If specimen cannot be processed in <1 hour, If only one tube, inoculate into Tube microbiology Tube T-I medium. 1 is a priority 3 For details consult Lab Manual or Standard Tube Operating Procedures Chemical analysis: 2 Record overall appearance; protein and glucose tests perform cell count: WBCs Microbiological tests PCR Immunochromatographic Centrifuge Latex Test Agglutination Freeze 250µl-500µl Test of CSF for PCR 100-200µl CSF volume is needed Do not centrifuge if<1ml (if < 500µl CSF When using kits refer When using kits refer to available, freeze to manufacturers manufacturers standard any remaining CSF standard operating procedures (SOPs) operating procedures (SOPs) volume for PCR) in package insert ( Supernatant in package insert Pellet ( Gram Stain Culture on Blood Agar Plate (BAP) and Chocolate Agar Plate (CAP) Decolorize with 95% ethanol; do not decolorize longer than 10 sec. Use sheep or horse blood, NOT human blood S. pneumoniae: H. influenzae: N. meningitidis: BAP CAP BAP or CAP LOOK FOR: gram POS. LOOK FOR: gram NEG. LOOK FOR: gram NEG. diplococci or cocci coccobacilli coffee-bean in short chains shaped diplococci S. pneumoniae H. influenzae N. meningitidis FOR anY otheR GRam stain Results, LOOK FOR: LOOK FOR: LOOK FOR: applY otheR Routine lab methods: Notify clinician of Gram small, grayish, moist (sometimes large, round, colorless- -

Streptococci
STREPTOCOCCI Streptococci are Gram-positive, nonmotile, nonsporeforming, catalase-negative cocci that occur in pairs or chains. Older cultures may lose their Gram-positive character. Most streptococci are facultative anaerobes, and some are obligate (strict) anaerobes. Most require enriched media (blood agar). Streptococci are subdivided into groups by antibodies that recognize surface antigens (Fig. 11). These groups may include one or more species. Serologic grouping is based on antigenic differences in cell wall carbohydrates (groups A to V), in cell wall pili-associated protein, and in the polysaccharide capsule in group B streptococci. Rebecca Lancefield developed the serologic classification scheme in 1933. β-hemolytic strains possess group-specific cell wall antigens, most of which are carbohydrates. These antigens can be detected by immunologic assays and have been useful for the rapid identification of some important streptococcal pathogens. The most important groupable streptococci are A, B and D. Among the groupable streptococci, infectious disease (particularly pharyngitis) is caused by group A. Group A streptococci have a hyaluronic acid capsule. Streptococcus pneumoniae (a major cause of human pneumonia) and Streptococcus mutans and other so-called viridans streptococci (among the causes of dental caries) do not possess group antigen. Streptococcus pneumoniae has a polysaccharide capsule that acts as a virulence factor for the organism; more than 90 different serotypes are known, and these types differ in virulence. Fig. 1 Streptococci - clasiffication. Group A streptococci causes: Strep throat - a sore, red throat, sometimes with white spots on the tonsils Scarlet fever - an illness that follows strep throat. It causes a red rash on the body. -

High Frequency of Multidrug Resistant Urinary Isolates in Pregnant
www.symbiosisonline.org Symbiosis www.symbiosisonlinepublishing.com Research Article SOJ Microbiology & Infectious Diseases Open Access High Frequency of Multidrug Resistant Urinary Isolates in Pregnant Women in a Tertiary Care Hospital of Nepal Bhandari Pashupati1*, Dipak Raj Joshi2, Sharma Khem Raj2, Khanal Santosh1, Acharya Ganesh3, Adhikari Nabaraj2 1Department of Microbiology, National College (NIST), Khusibu, Kathmandu, Nepal 2Department of Microbiology, Kantipur College of Medical Sciences, Sitapaila, Kathmandu, Nepal 3Department of Pathology Laboratory, Paropakar Maternity and Women’s Hospital, Kathmandu, Nepal Received: November 15, 2016; Accepted: December 25, 2016; Published: January 2, 2017 *Corresponding author: Bhandari Pashupati, Department of Microbiology, National College (NIST), Khusibu, Kathmandu, Nepal; E-mail: pa- [email protected] Introduction Abstract Urinary Tract Infection (UTI) is one of the most common Introduction: Urinary tract infection (UTI) is a common infection during pregnancy with frequency ranging from 5-20%. bacterial infections accounting for about 25% of all types of Bacteriuria in pregnancy whether asymptomatic or symptomatic if infection in human and approximately 10% of the humans untreated may lead to pyelonephritis which may result in abortion, acquire UTI at some time during their lifetime [1]. The Incidence premature delivery, low weight birth and even still birth. This study of UTI is age and sex dependent and it occurs with higher was conducted to identify the etiological agents and assess antibiotic frequency in female than in men because of shorter urethra and resistance pattern of bacterial urinary isolates in patients attending close proximity of urinary tract to anus and most prevalent age Paropakar Maternity and Women’s Hospital, Kathmandu, Nepal. group experiencing UTI in females is 21–30 years age [2]. -

Branda Jcm 2013.Pdf
JCM Accepts, published online ahead of print on 19 June 2013 J. Clin. Microbiol. doi:10.1128/JCM.00824-13 Copyright © 2013, American Society for Microbiology. All Rights Reserved. 1 Performance of the VITEK MS v2.0 system in distinguishing 2 Streptococcus pneumoniae from non-pneumococcal species of the 3 Streptococcus mitis group 4 5 6 John A. Branda1,2#, Rachelle P. Markham1, Cherilyn D. Garner3, Jenna A. Rychert1,2 and 7 Mary Jane Ferraro1,2 8 DISTRIBUTE 9 10 Department of Pathology, 1Massachusetts General Hospital and 2Harvard Medical 11 School, Boston, Massachusetts OR 12 3R&D Microbiology, bioMérieux, Inc., Hazelwood, MO 13 14 Running title: Pneumococcal vs non-pneumococcal strep using mass spec 15 16 #Corresponding author address: COPY 17 55 Fruit St., GRB-526 18 Boston, MA 02114-2696 NOT 19 Telephone: 617-726-3611 20 Fax: 617-726-5957 DO 21 Email: [email protected] 22 ONLY REVIEW 1 FOR 23 Abstract 24 The VITEK MS v2.0 MALDI-TOF mass spectrometry system accurately distinguished S. 25 pneumoniae from non-pneumococcal S. mitis group species. Only 1 of 116 non- 26 pneumococcal isolates (<1%) was misidentified as S. pneumoniae. None of 95 27 pneumococcal isolates was misidentified. This method provides a rapid, simple means of 28 discriminating among these challenging organisms. DISTRIBUTE 29 30 Short-form paper OR 31 Using conventional phenotypic identification methods, it has been challenging for 32 clinical laboratories to distinguish accurately between bacterial species within certain 33 groups, such as the coagulase-negative staphylococci or COPYthe nonfermenting gram-negative 34 bacilli. The Streptococcus mitis group is another set of closely-related species between 35 which conventional identification methods cannot reliably differentiate. -

S.Pneumoniae the New VITEK® 2 ■ S.Pneumoniae? in the Early Years of Antibiotherapy Identifying System Resistance ■ What Are the Key with Penicillin
r te et sl 5 ew 00 l N 2 na ry tio ua na br ter Fe In 5 • n° State-of-the-Art The bioMérieux Did you know? Practical advice Streptococcus pneumoniae solution has always been an important pathogen ■ ■ ■ ■ for man and a big therapeutic success Antibiotic and Evaluation of Web sites why test S.pneumoniae the New VITEK® 2 ■ S.pneumoniae? in the early years of antibiotherapy Identifying System Resistance ■ what are the key with penicillin. News issues with S.pneumoniae For many years ■ NCCLS infections? this species was perfectly susceptible recommendations ■ What is the and antibiotic susceptibility testing ■ Antibiotics reference method for testing ■ ® was not necessary. VITEK 2 S.pneumoniae? phenotypes Then things changed dramatically about ■ The epidemiology of 25 years ago and resistance emerged S.pneumoniae in the South Hemisphere, spread to the world and now amounts State-of-the-Art to 50% of strains for penicillin as well as for other antibiotic families. This is a perfect example of emergence Antibiotic and of new resistance. Antibiotic testing of this species S.pneumoniae is now highly important and bioMérieux has developed a special VITEK2 card Keith Klugman MB BCh, PhD, is Professor of International Health at the Rollins School of Public Health at Emory University, in Atlanta, GA, USA. He is also Professor of Medicine in for this purpose with an appropriate the Division of Infectious Diseases of the School of Medicine at that University and a selection of antibiotics. Visiting Researcher in the Respiratory Diseases Branch of the Centers for Disease Control and Prevention (CDC). -

Abnormal Vaginal Microflora: Risk Factors
RĪGA STRADIŅŠ UNIVERSITY Jana Žodžika ABNORMAL VAGINAL MICROFLORA: RISK FACTORS, BED-SIDE DIAGNOSTIC METHODS IN PREGNANCY AND EFFICIENCY OF AN ALTERNATIVE NON-ANTIBACTERIAL TREATMENT MODALITY IN PREGNANT AND NON-PREGNANT WOMEN For obtaining the degree of a Doctor of Medicine Speciality Obstetrics and Gynaecology Research supervisor: MD, PhD, Professor Dace Rezeberga, Rīga Stradiņš University (Latvia) Research scientific consultant: MD, PhD, Professor Gilbert Donders, University of Antwerp (Belgium) Riga, 2014 ANNOTATION Normal vaginal microflora is an important women`s health factor, maintained by high numbers of different Lactobacillus species. Abnormal vaginal microflora and infections ascending from the lower urogenital tract represent an important reason for abortions, preterm delivery and neonatal infections. Multiple investigators have attempted to identify the patients at risk for preterm labor, followed by the treatment in a low risk population of genital infections, but the results did not meet initial hopes. Still there is growing evidence that treatment of abnormal vaginal microflora with adequate antibiotics in early pregnancy can prevent at least some of the infections related to preterm birth. While antimicrobial agents cure infections, they can cause side effects. Furthermore, urogenital pathogen drug resistance is on the increase and disrupt protective vaginal microflora. Many pregnant women are very anxious about taking antibiotics because of potentially adverse effects on the newborn. During pregnancy treatment that restores normal vaginal flora and acidity without systemic effects would be preferable to any other treatment. The aim of the present study is to investigate the influence of vaginal application of ascorbic acid (vitamin C) on abnormal vaginal microflora during pregnancy, and also to identify risk group and assess the validity of the “bed-side” diagnostic tests during the first antenatal visit in order to detect different types of abnormal vaginal flora. -

Optochin Test
UK Standards for Microbiology Investigations Optochin test Issued by the Standards Unit, National Infection Service, PHE Bacteriology – Test Procedures | TP 25 | Issue no: 4 | Issue date: 03.12.18 | Page: 1 of 13 © Crown copyright 2018 Optochin test Acknowledgments UK Standards for Microbiology Investigations (SMIs) are developed under the auspices of Public Health England (PHE) working in partnership with the National Health Service (NHS), Public Health Wales and with the professional organisations whose logos are displayed below and listed on the website https://www.gov.uk/uk- standards-for-microbiology-investigations-smi-quality-and-consistency-in-clinical- laboratories. SMIs are developed, reviewed and revised by various working groups which are overseen by a steering committee (see https://www.gov.uk/government/groups/standards-for-microbiology-investigations- steering-committee). The contributions of many individuals in clinical, specialist and reference laboratories who have provided information and comments during the development of this document are acknowledged. We are grateful to the medical editors for editing the medical content. For further information please contact us at: Standards Unit National Infection Service Public Health England 61 Colindale Avenue London NW9 5EQ E-mail: [email protected] Website: https://www.gov.uk/uk-standards-for-microbiology-investigations-smi-quality- and-consistency-in-clinical-laboratories PHE publications gateway number: 2018381 UK Standards for Microbiology Investigations are produced in association with: Logos correct at time of publishing. Bacteriology – Test Procedures | TP 25 | Issue no: 4 | Issue date: 03.12.18 | Page: 2 of 13 UK Standards for Microbiology Investigations | Issued by the Standards Unit, Public Health England Optochin test Contents Acknowledgments ................................................................................................................