Incucyte™ ZOOM User Manual 2016A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

E-37-V Dome Camera Operation Manual
E-37-V Dome Camera Operation Manual i Table of Contents 1 Network Config .............................................................................................................. 1 1.1 Network Connection .......................................................................................... 1 1.2 Log in ................................................................................................................ 1 2 Live ................................................................................................................................ 6 2.1 Encode Setup ................................................................................................... 6 2.2 System Menu .................................................................................................... 7 2.3 Video Window Function Option ......................................................................... 7 2.4 Video Window Setup ......................................................................................... 8 2.4.1 Image Adjustment ....................................................................................... 8 2.4.2 Original Size ............................................................................................... 9 2.4.3 Full Screen ................................................................................................. 9 2.4.4 Width and Height Ratio ............................................................................. 10 2.4.5 Fluency Adjustment ................................................................................. -

Class -IV Super Computer Year- 2020-21
s Class -IV Super Computer Year- 2020-21 2. Windows 7 ❖ Focus of the Chapter 1. Windows desktop 2. Desktop icons 3. Start Menu 4. Task bar 5. Files and folders 6. Creating & saving new file/folder 7. Selecting a file/folder 8. Opening a file/folder 9. Renaming a file/folder 10. Deleting a file/folder 11. Moving a file/folder 12. Copying a file/folder 13. Creating a shortcut to a file/folder Keywords • Booting – Loading of the operating system. • Taskbar- The long bar present at the bottom of the desktop • Notification area- The area located on the right side of the taskbar • Folder- A container for storing files and other folders. Introduction Windows 7 is an operating system that Microsoft has produced for use on personal computers. It is the follow-up to the Windows Vista Operating System, which was released in 2006. An operating system allows your computer to manage software and perform essential tasks. It is also a Graphical User Interface (GUI) that allows you to visually interact with your computer’s functions in a logical, fun, and easy way. Interact with your computer’s functions in a logical, fun, and easy way. * The first screen appear after you turn on the power of computer is a desktop • If it is a shared PC; more than one user uses it, or one user with password protected, you will arrive at Welcome Screen Desktop Components 1- Icons: An icon is a graphic image, a small picture or object that represents a file, program, web page, or command. -
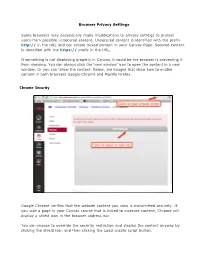
Chrome Security
Browser Privacy Settings Some browsers may occasionally make modifications to privacy settings to protect users from possible unsecured content. Unsecured content is identified with the prefix http:// in the URL and can create mixed content in your Canvas Page. Secured content is identified with the https:// prefix in the URL. If something is not displaying properly in Canvas, it could be the browser is preventing it from showing. You can always click the ‘new window’ icon to open the content in a new window. Or you can ‘allow the content. Below, are images that show how to enable content in both browsers Google Chrome and Mozilla Firefox. Chrome Security Google Chrome verifies that the website content you view is transmitted securely. If you visit a page in your Canvas course that is linked to insecure content, Chrome will display a shield icon in the browser address bar. You can choose to override the security restriction and display the content anyway by clicking the shield icon and then clicking the Load unsafe script button. Chrome Media Permissions Chrome has its own media permission within the browser. To use your computer camera and microphone within any Canvas feature, you will have to approve two permissions: 1. Allow access to Canvas via the Adobe Flash Player Settings. This prompt appears in the center of the video and audio pop-up windows, or the center of the browser if you are accessing Conversations. 2. Allow access to Canvas via Chrome's media permission settings. This prompt appears just below the address bar. Click the Allow button. -

CUSTOMIZING YOUR TOOLBARS in INTERNET EXPLORER, OUTLOOK EXPRESS, WINDOWS and WORD a Seminar Presented by Bill Wilkinson April 2008
CUSTOMIZING YOUR TOOLBARS IN INTERNET EXPLORER, OUTLOOK EXPRESS, WINDOWS AND WORD A Seminar Presented by Bill Wilkinson April 2008 Internet Explorer 7 Toolbars Internet Explorer 7, the latest version of Microsoft’s Web browser, sports a cleaner look than its predecessor. In fact, some users might wonder where everything went. The main toolbar is reduced in IE7 to just a couple of main buttons in the upper-left corner near the Address bar and a smattering of smaller buttons scattered about. The Menu Bar, a familiar feature in just about every program, was hidden by default in the earliest version IE7, but Microsoft was shamed into returning it to its traditional place. If you downloaded IE7 when it first came out, you will not see the Menu Bar. We will show you how to access it during this seminar session. The IE7 standard install also lacked a visible Links toolbar. Fortunately, Internet Explorer 7 features a streamlined toolbar that you can customize if you find it a little too sparse. The Page button is really a drop-down menu of commands loosely related to viewing the page in your browser window. This is where you can change text size, zoom level, or send pages and links through email. The Copy and Paste commands also ended up here, along with the command for opening a new window . Clicking Tools opens another submenu with a lot of the same commands from the original Tools menu. Here you’ll find the Pop-up Blocker, the Internet Options command, and other settings. Rounding out the IE7 interface are star-shaped buttons on the left side of the screen to add and view Favorites (called the Favorites Center). -

Poly Video Mode Administrator Guide
ADMINISTRATOR GUIDE 3.0.0 | December 2019 | 3725-85854-004A Poly Video Mode Getting Help For more information about installing, configuring, and administering Poly/Polycom products or services, go to Polycom Support. Plantronics, Inc. (Poly — formerly Plantronics and Polycom) 345 Encinal Street Santa Cruz, California 95060 © 2019 Plantronics, Inc. All rights reserved. Poly, the propeller design, and the Poly logo are trademarks of Plantronics, Inc. All other trademarks are the property of their respective owners. Contents Before You Begin................................................................................................6 Audience, Purpose, and Required Skills.............................................................................6 Related Poly and Partner Resources..................................................................................6 Getting Started................................................................................................... 8 Product Overview of Poly Video Systems...........................................................................8 Administrator Features and Capabilities.................................................................. 9 Powering the System On and Off......................................................................................10 Navigating the System...................................................................................................... 10 Access the System Web Interface..........................................................................10 -

Internet Explorer 11
® Microsoft Internet Explorer 11 Qu ick Reference Card Internet Explorer 11 Window Keyboard Shortcuts Address Bar list arrow Security report General Search Refresh View favorites, feeds and history New Window <Ctrl> + <N> Navigation Home New Tab <Ctrl> + <T> Buttons Address Tab New tab Close Window/Tab <Ctrl> + <W> Bar Open link in New Tab <Ctrl> + <Shift> Tools (Foreground) + click Open link in New Tab <Ctrl> + click (Background) Select Address Bar <Alt> + <D> Find <Ctrl> + <F> Print <Ctrl> + <P> Select All <Ctrl> + <A> Zoom In <Ctrl> + <+> Zoom Out <Ctrl> + <-> Full Screen Mode <F11> Help <F1> Add Favorite <Ctrl> + <D> Refresh <F5> Stop <Esc> Navigation Next item <Tab> Up one screen <Page Up> Scroll Bars Down one screen <Page Down> Home <Alt> + <Home> The Fundamentals Tools <Alt> + <X> Favorites <Alt> + <C> • A web address, also known as a URL (Uniform • To Search the Web: Click the Search button in the Resource Locator) is made up of two main parts: Address Bar, type a keyword or term in the Address Bar View Address Bar <F4> ü http:// (Hypertext Transfer Protocol): the set of and press <Enter>. History rules for exchanging files on the World Wide • To Change the Search Provider: Click the Address Bar Go Forward <Alt> + <à> Web. list arrow in the Address bar and select the provider Go Back <Alt> + <ß> ü Domain name: the unique name that you wish to use from the row of icons along the bottom of Switch to previous Tab <Ctrl> + <Shift> identifies a specific site on the web. the menu. + <Tab> • To Add a Search Provider: Click the Address Bar list • To Open a Web Page: Click the Address Bar and Switch to next Tab <Ctrl> + <Tab> type the address of the Web page. -

Chromebook Cheat Sheet Quick Tips to Keep You Rockin’!
Chromebook Cheat Sheet Quick Tips to keep you rockin’! 1) Power 2) Connecting to the Internet 3) Logging in 4) Touchpad Navigation 1 finger on touchpad 2 fingers on touchpad at the same time 3 fingers on touchpad at the same time 5) Keyboard CAPS LOCK Print Screen 6) “Top 10” Chromebook Keyboard Shortcuts 10. Take a Screenshot on a Chromebook 9. Open File Manager 8. Open Task Manager 7. Launch Apps on the Shelf 6. Lock the Screen 5. Open the Hotdog 4. Toggle Caps Lock On/Off 3. Logout 2. Mirror Monitor 1. Show Keyboard Shortcut Overlay 7) A bunch more keyboard shortcuts 1) Power Opening the clamshell lid of a Chromebook will turn it on. If it’s already open but powered off, push the power button in the upper-right corner of the keyboard. Be aware that just closing the lid does NOT turn it off, and will slowly drain the battery. To power the Chromebook down, either use the “Shut down” button in the lower-right menu, or push and hold the Power button at the upper-right of the keyboard. 2) Connecting to the Internet Chromebooks are basically just internet browsing devices. If you start up a school Chromebook and get the dinosaur screen - “Can’t connect to internet” - turning it off and turning it back on will usually get it to connect. 3) Logging in & out Chromebooks require a slightly different log on than “regular” computers. Users need to enter their entire school email address (not just initial and last name) and their regular school password. -

Google Chrome Settings
Google Chrome Settings Opening the Settings Page You can open the Settings page by clicking on the icon with three stacked horizontal lines to the left of the address bar; this will open up a dropdown menu, and Settings will be located to the bottom of the screen. Users can also type in chrome://chrome/settings/ into the address bar to locate the page, and Mac OS X users can also open the Settings page by choosing Chrome > PreferencesBrowser Settings or hit ⌘, (Command key plus the comma key) 1. 2. Opena. theNote Settings – For mostpage (directionsusers they willabove) not see this setting until they expose Locate the “Privacy” section and choose “Content Settings…” clickadvanced on it settings; have the user scroll to the bottom of the page, and 3. In the “Contentif there is set a link that says “Show advance settings…” ask the user to tings” overlay ensure that the following options are set: a. Cookies b. Cookies - unchecked: Allow local data to be set (recommended) c. JavaScript: The option for “Block third party cookies and data” is d. Pop-ups all sites to show pop-ups i. Note: Allow If the all user sites does to run not JavaScript want to enable (recommended) popups for all sites, : Allow open up: a “Pop- thehave URL them of thego click online on bankingthe “Manage domain exceptions…” and set th button to up exceptions” overlay; here have them type e Behavior to Security Allow 1. 2. Opena. theNote Settings – For mostpage (directionsusers they willabove) not see this setting until they expose Locate the “HTTPS/SSL” section and choose “Content Settings…”m of the page, and clickadvanced on it settings; have the user scroll to the botto 3. -

CSC245 - Web Technologies and Programming Homework Assignment 5: Baby Names
CSC245 - Web Technologies and Programming Homework Assignment 5: Baby Names This assignment tests your understanding of fetching data from files and web services using Ajax requests. You must match in appearance and behavior the following web page: Background Information: Every 10 years, the Social Security Administration provides data about the 1000 most popular boy and girl names for children born in the US, at http://www.ssa.gov/OACT/babynames/ . Your task for this assignment is to write the JavaScript code for a web page to display the baby names, popularity rankings, and meanings. You do not need to submit any HTML or CSS file. You will be provided with the HTML (names.html) and CSS (names.css) code to use. Turn in only the following file: ◦ names.Js, the JavaScript code for your Baby Names web page Data: Your program will get its data from a web service located at the following URL: https://cs.berry.edu/webdev/scripts/labs/babynames/babynames.php This web service has three “modes,” each of which provides a different type of data. Specify which type of data you want from it by changing a query parameter named type. Each query returns either plain text or JSON as its output. If you make a malformed request, such as one missing a necessary parameter, the service will respond with an HTTP error code of 400 rather than the default 200. If you request data for a name the server doesn't have data for, the service will respond with an error code of 404. (Hint: You can test queries by typing the URL of the web service, along with the appropriate query string parameters, in your web browser's address bar and seeing the result. -

Site Hyperlinks Help Card
Site Hyperlinks Help Card Creating and Using Links You can insert hyperlinks to external web sites as well as to other pages of your web site Use the Hyperlink Manager to create links to other web sites and to other pages on your site. The Page Link tool can also be used for linking pages. Configure your links to open up in a new browser window, so that users do not navigate away from the page they were on Hyperlinks to external web sites Hyperlinks to other Site Pages Hyperlinks to other Site Pages (Using Page Link tool) 1. Position your cursor at the location 1. Navigate to the page you want to link to 1. Position your cursor at the location where you where you want to insert the link 2. Copy the page address from the want to insert the link on the page and click the Hyperlink Manager address bar of your browser into a 2. Click the Page Link icon notepad 3. Locate the name of the Page you want to link 2. In the URL field, type in the 3. Position your cursor at the location to. You will need to use the “1 2 3 4..” address of the web site you want where you want to insert the link on navigation links to move from one screen to the to link to the page next as shown below: 3. Type the text you want to display 4. Click the Hyperlink Manager icon as the link into the Link Text field 5. In the URL field, paste in the address 4. -

Allowing Popups for Xpressdox.Com – for Chrome, Edge, Firefox (Allows the Preview Button in Xpressdox to Function Properly)
Allowing Popups For Xpressdox.com – For Chrome, Edge, Firefox (Allows the Preview button in XpressDox to function properly) Chrome (version 88+): You could click on the icon in your address bar to allow the popups from this site. Simply click on this icon and select the allow option: Additionally, you could follow these steps: 1. On your computer, open Chrome. 2. At the top right, click More . 3. Click Settings. 4. Under 'Privacy and security', click Site settings. 5. Click Pop-ups and redirects. (towards the bottom of the listing) 6. Under 'Allow', look for the site (iowadocs.xpressdox.com). NOTE: if this entry is here try removing this and adding it back in. 7. Click “Add” button beside “Allow” and add [*.]xpressdox.com. (* means it will allow all pop ups from this domain) Edge (version 88+): You could click on the icon in your address bar to allow the popups from this site . Simply click on this icon and select the allow option: Additionally, you could follow these steps: 1. On your computer, open Edge. 2. At the top right, click “Setting and more…” button (or click Alt + F). 3. Click Settings . 4. Click “Cookies and site permissions”. 5. Find “All permissions” section. 6. Click on “Pop-ups and redirects”. 7. Under “Allow” click on the “Add” button. 8. Add [*.]xpressdox.com. (* means it will allow all pop ups from this domain) FAQ_Allowing Popups For Xpressdox.docx Page 1 of 2 Firefox (version 85+): When blocking a pop-up, Firefox displays an information bar (if it hasn’t been previously dismissed – see below), as well as an icon in the address bar. -

Restart the Computer
Gloucester County Library System COMPUTER BASICS WINDOWS XP VISTA, WINDOW 7 Computer Classes Check the GCLS online calendar for the schedule of Computer Classes www.gcls.org Basic Computer Skills: Required for all other computer classes. Learn how to use the mouse, open and close programs, select items and text. Internet Basics: Learn how to use the Internet, click links, navigate sites and print useful information. Email Basics: Learn about email, create your own email address and get some valuable practice. Software Basics: Overview of common office software such as Microsoft Word, Excel and PowerPoint. When Caring for a PC Good Practices Plug your PC, monitor, printer, scanner, etc. into a surge protector Keep your PC in a well-ventilated area Keep your PC away from moisture, and moisture away from your PC Properly turn off your PC and monitor when not in use • Safety Tips Sit in a comfortable chair when using a PC Make sure your PC is on a stable surface NEVER open the case on your PC while it is plugged in Take breaks often to rest your eyes and increase circulation • Anti-Virus Software Purchase anti-virus software and keep it up-to-date Scan your PC each time you turn on your PC Anti-virus software will protect your computer from dangerous viruses • Basic Cleaning Clean your monitor with a clean soft cloth using water or isopropyl alcohol if needed Clean your keyboard with a clean soft cloth – if needed, use isopropyl alcohol to remove grime – only use a small amount on a clean soft cloth.