2016 Project Abstract for the Period Ending June 30, 2019
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hemlock Ravine Scientific and Natural Area 1984 Resource Inventory
This document is made available electronically by the Minnesota Legislative Reference Library as part of an ongoing digital archiving project. http://www.leg.state.mn.us/lrl/lrl.asp Hemlock Ravine Scientific and Natural Area 1984 Resource Inventory Portions of SE 1/4, Section 3 Township 48, Range 16W Esko Quadrangle - L20a Carlton County Minnesota Prepared by The Scientific and Natural Area Program and The Minnesota Natural Heritage Program Division of Fish and Wildlife Minnesota Department of Natural Resources June 1984 Scientific and Natural Areas Scientific and Natural Areas serve: Education - elementary through high school groups use such areas as outdoor classrooms. Nature Observation - the public uses these areas to observe Minnesota's most unique or rare natural resources. Protection Functions - Minnesota's rarest species or most unique features are protected for the citizens of today and tomorro,,;·]. Recreation - the public uses such areas for informal, dispersed recreation 0 Resea~ch - colleges are able to establish long term research projects secure in knoT:!ing the area will not be influenced by other management activities. Genetic Storehouse - ?otentially valuable plants ,and animals are retained thereby ·offering potential for new medicines, resistance to plant diseases, and other unknown secrets. Currently there are 34 Scientific and Natural Areas protecting undisturbed remnants of Minnesota's plant communities and plant and animal species. These areas encompass maple basswood forests, virgin prairies, orchid bogs, heron rookeries, sand dunes, and virgin pine stands, as wel~ as many rare plant and animal species. o Y4 % mile I· I• , • I • I o 200m 400m 800m HEMLOCK RAVI ES A VICINITY o ~ % mile I. -

Minnesota Statutes 2020, Chapter 85
1 MINNESOTA STATUTES 2020 85.011 CHAPTER 85 DIVISION OF PARKS AND RECREATION STATE PARKS, RECREATION AREAS, AND WAYSIDES 85.06 SCHOOLHOUSES IN CERTAIN STATE PARKS. 85.011 CONFIRMATION OF CREATION AND 85.20 VIOLATIONS OF RULES; LITTERING; PENALTIES. ESTABLISHMENT OF STATE PARKS, STATE 85.205 RECEPTACLES FOR RECYCLING. RECREATION AREAS, AND WAYSIDES. 85.21 STATE OPERATION OF PARK, MONUMENT, 85.0115 NOTICE OF ADDITIONS AND DELETIONS. RECREATION AREA AND WAYSIDE FACILITIES; 85.012 STATE PARKS. LICENSE NOT REQUIRED. 85.013 STATE RECREATION AREAS AND WAYSIDES. 85.22 STATE PARKS WORKING CAPITAL ACCOUNT. 85.014 PRIOR LAWS NOT ALTERED; REVISOR'S DUTIES. 85.23 COOPERATIVE LEASES OF AGRICULTURAL 85.0145 ACQUIRING LAND FOR FACILITIES. LANDS. 85.0146 CUYUNA COUNTRY STATE RECREATION AREA; 85.32 STATE WATER TRAILS. CITIZENS ADVISORY COUNCIL. 85.33 ST. CROIX WILD RIVER AREA; LIMITATIONS ON STATE TRAILS POWER BOATING. 85.015 STATE TRAILS. 85.34 FORT SNELLING LEASE. 85.0155 LAKE SUPERIOR WATER TRAIL. TRAIL PASSES 85.0156 MISSISSIPPI WHITEWATER TRAIL. 85.40 DEFINITIONS. 85.016 BICYCLE TRAIL PROGRAM. 85.41 CROSS-COUNTRY-SKI PASSES. 85.017 TRAIL REGISTRY. 85.42 USER FEE; VALIDITY. 85.018 TRAIL USE; VEHICLES REGULATED, RESTRICTED. 85.43 DISPOSITION OF RECEIPTS; PURPOSE. ADMINISTRATION 85.44 CROSS-COUNTRY-SKI TRAIL GRANT-IN-AID 85.019 LOCAL RECREATION GRANTS. PROGRAM. 85.021 ACQUIRING LAND; MINNESOTA VALLEY TRAIL. 85.45 PENALTIES. 85.04 ENFORCEMENT DIVISION EMPLOYEES. 85.46 HORSE -

The Campground Host Volunteer Program
CAMPGROUND HOST PROGRAM THE CAMPGROUND HOST VOLUNTEER PROGRAM MINNESOTA DEPARTMENT OF NATURAL RESOURCES 1 CAMPGROUND HOST PROGRAM DIVISION OF PARKS AND RECREATION Introduction This packet is designed to give you the information necessary to apply for a campground host position. Applications will be accepted all year but must be received at least 30 days in advance of the time you wish to serve as a host. Please send completed applications to the park manager for the park or forest campground in which you are interested. Addresses are listed at the back of this brochure. General questions and inquiries may be directed to: Campground Host Coordinator DNR-Parks and Recreation 500 Lafayette Road St. Paul, MN 55155-4039 651-259-5607 [email protected] Principal Duties and Responsibilities During the period from May to October, the volunteer serves as a "live in" host at a state park or state forest campground for at least a four-week period. The primary responsibility is to assist campers by answering questions and explaining campground rules in a cheerful and helpful manner. Campground Host volunteers should be familiar with state park and forest campground rules and should become familiar with local points of interest and the location where local services can be obtained. Volunteers perform light maintenance work around the campground such as litter pickup, sweeping, stocking supplies in toilet buildings and making emergency minor repairs when possible. Campground Host volunteers may be requested to assist in the naturalist program by posting and distributing schedules, publicizing programs or helping with programs. Volunteers will set an example by being model campers, practicing good housekeeping at all times in and around the host site, and by observing all rules. -

Minnesota State Parks.Pdf
Table of Contents 1. Afton State Park 4 2. Banning State Park 6 3. Bear Head Lake State Park 8 4. Beaver Creek Valley State Park 10 5. Big Bog State Park 12 6. Big Stone Lake State Park 14 7. Blue Mounds State Park 16 8. Buffalo River State Park 18 9. Camden State Park 20 10. Carley State Park 22 11. Cascade River State Park 24 12. Charles A. Lindbergh State Park 26 13. Crow Wing State Park 28 14. Cuyuna Country State Park 30 15. Father Hennepin State Park 32 16. Flandrau State Park 34 17. Forestville/Mystery Cave State Park 36 18. Fort Ridgely State Park 38 19. Fort Snelling State Park 40 20. Franz Jevne State Park 42 21. Frontenac State Park 44 22. George H. Crosby Manitou State Park 46 23. Glacial Lakes State Park 48 24. Glendalough State Park 50 25. Gooseberry Falls State Park 52 26. Grand Portage State Park 54 27. Great River Bluffs State Park 56 28. Hayes Lake State Park 58 29. Hill Annex Mine State Park 60 30. Interstate State Park 62 31. Itasca State Park 64 32. Jay Cooke State Park 66 33. John A. Latsch State Park 68 34. Judge C.R. Magney State Park 70 1 35. Kilen Woods State Park 72 36. Lac qui Parle State Park 74 37. Lake Bemidji State Park 76 38. Lake Bronson State Park 78 39. Lake Carlos State Park 80 40. Lake Louise State Park 82 41. Lake Maria State Park 84 42. Lake Shetek State Park 86 43. -

Kettle River, Minnesota
Kettle River, Minnesota 1. The region surrounding the river: a. The Kettle River is located in east-central Minnesota. The river has its headwaters in Carlton County and flows generally north-south, passing through Pine County and into the St. Croix River. The basin has a long history of faults and glacial activity. The bedrock formations are of pre-Cambrian metamorphic and volcanic rock. This layer is covered by Cambrian sandstone and unconsolidated glacial till. Outcroppings of sandstone and pre-Cambrian lava are frequent. The area is ragged and rolling with dramatic local relief. The area has gone through a dramatic ecological change since the logging days when the white pine was the dominant vegetation. Today the region has a varied pattern of red pine, spruce, white pine, white birch maple, oak, aspen, and basswood. Major transportation lines in the area include Interstate 35 running north-south through the basin and Minnesota 23 running northeast- southwest through the basin. Minnesota 48 crosses the river east-west just east of Hinckley, Minnesota, and Minnesota Route 65 runs north-south about 25 miles west of the river. Land use in the basin is limited to agriculture and timber production. The Mhmeapolis-St. Paul area to the south supports heavy industry and manufacturing. b. Population within a 50-mile radius was estimated at 150, 700 in 1970. The Duluth, Minnesota/Superior, Wisconsin, metropolitan area lies just outside the 50-mile radius and had an additional 132, 800 persons in 1970. c. Numerous state forests are found in this part of Minnesota. They are Chengwatona State Forest, DAR State Forest, General C. -

Northwest Lowlands Ecological Landscape
Chapter 16 Northwest Lowlands Ecological Landscape Where to Find the Publication The Ecological Landscapes of Wisconsin publication is available online, in CD format, and in limited quantities as a hard copy. Individual chapters are available for download in PDF format through the Wisconsin DNR website (http://dnr.wi.gov/, keyword “landscapes”). The introductory chapters (Part 1) and supporting materials (Part 3) should be downloaded along with individual ecological landscape chapters in Part 2 to aid in understanding and using the ecological landscape chapters. In addition to containing the full chapter of each ecological landscape, the website highlights key information such as the ecological landscape at a glance, Species of Greatest Conservation Need, natural community management opportunities, general management opportunities, and ecological landscape and Landtype Association maps (Appendix K of each ecological landscape chapter). These web pages are meant to be dynamic and were designed to work in close association with materials from the Wisconsin Wildlife Action Plan as well as with information on Wisconsin’s natural communities from the Wisconsin Natural Heritage Inventory Program. If you have a need for a CD or paper copy of this book, you may request one from Dreux Watermolen, Wisconsin Department of Natural Resources, P.O. Box 7921, Madison, WI 53707. Photos (L to R): Red-shouldered Hawk, photo © Laurie Smaglick Johnson; arctic fritillary, photo by Ann Thering; Sedge Wren, photo © Laurie Smaglick Johnson; gray wolf, photo by Gary Cramer, U.S. Fish and Wildlife Service; Golden-winged Warbler, photo © Laurie Smaglick Johnson. Suggested Citation Wisconsin Department of Natural Resources. 2015. The ecological landscapes of Wisconsin: An assessment of ecological resources and a guide to planning sustainable management. -

RV Sites in the United States Location Map 110-Mile Park Map 35 Mile
RV sites in the United States This GPS POI file is available here: https://poidirectory.com/poifiles/united_states/accommodation/RV_MH-US.html Location Map 110-Mile Park Map 35 Mile Camp Map 370 Lakeside Park Map 5 Star RV Map 566 Piney Creek Horse Camp Map 7 Oaks RV Park Map 8th and Bridge RV Map A AAA RV Map A and A Mesa Verde RV Map A H Hogue Map A H Stephens Historic Park Map A J Jolly County Park Map A Mountain Top RV Map A-Bar-A RV/CG Map A. W. Jack Morgan County Par Map A.W. Marion State Park Map Abbeville RV Park Map Abbott Map Abbott Creek (Abbott Butte) Map Abilene State Park Map Abita Springs RV Resort (Oce Map Abram Rutt City Park Map Acadia National Parks Map Acadiana Park Map Ace RV Park Map Ackerman Map Ackley Creek Co Park Map Ackley Lake State Park Map Acorn East Map Acorn Valley Map Acorn West Map Ada Lake Map Adam County Fairgrounds Map Adams City CG Map Adams County Regional Park Map Adams Fork Map Page 1 Location Map Adams Grove Map Adelaide Map Adirondack Gateway Campgroun Map Admiralty RV and Resort Map Adolph Thomae Jr. County Par Map Adrian City CG Map Aerie Crag Map Aeroplane Mesa Map Afton Canyon Map Afton Landing Map Agate Beach Map Agnew Meadows Map Agricenter RV Park Map Agua Caliente County Park Map Agua Piedra Map Aguirre Spring Map Ahart Map Ahtanum State Forest Map Aiken State Park Map Aikens Creek West Map Ainsworth State Park Map Airplane Flat Map Airport Flat Map Airport Lake Park Map Airport Park Map Aitkin Co Campground Map Ajax Country Livin' I-49 RV Map Ajo Arena Map Ajo Community Golf Course Map -

Huntersville Forest Landing State Forest Campground
Off-Highway Vehicle Access to Huntersville Forest Landing State Forest Campground Frequently Asked Questions What is a state forest campground? State forest campgrounds are designated campgrounds in state forests. They provide basic needs while camping such as picnic tables, fire rings, water and toilets. Visitors should not typically expect showers, flush toilets or electric at campgrounds. The campgrounds are managed by the Department of Natural Resources Division of Parks and Trails. What is an off-highway vehicle? Off-highway vehicles (OHVs) consist of three classes: All-terrain vehicles (ATVs), off-road vehicles (ORVs) and off- highway motorcycles (OHMs). ATVs are three to six-wheeled recreational vehicles that are less than 65 inches in width. ORVs are recreational vehicles over 65 inches and OHMs are two-wheeled recreational vehicles. What uses are there currently at Huntersville Forest Landing Campground? Huntersville Forest Landing is managed as a general use campground. This campground has twenty-four drive-in campsites, including one handicap-accessible campsite, picnic tables, drinking water, and five vault toilets. The campground provides access to the Crow Wing River for swimming, fishing, and boating. The campground is considered "primitive," designed to furnish only the basic needs of the camper. The campsites consist of a cleared area, fire ring, and table. All sites are on a first-come, first-served basis. Why is the DNR doing this? Why Huntersville Forest Landing? The DNR recognizes that camping opportunities for campers with off-highway vehicles (OHVs) is limited in state forests. Allowing campers with OHVs direct access from campsites to trails will improve their recreational experience. -

Re-Thinking the Classification of Corticioid Fungi
mycological research 111 (2007) 1040–1063 journal homepage: www.elsevier.com/locate/mycres Re-thinking the classification of corticioid fungi Karl-Henrik LARSSON Go¨teborg University, Department of Plant and Environmental Sciences, Box 461, SE 405 30 Go¨teborg, Sweden article info abstract Article history: Corticioid fungi are basidiomycetes with effused basidiomata, a smooth, merulioid or Received 30 November 2005 hydnoid hymenophore, and holobasidia. These fungi used to be classified as a single Received in revised form family, Corticiaceae, but molecular phylogenetic analyses have shown that corticioid fungi 29 June 2007 are distributed among all major clades within Agaricomycetes. There is a relative consensus Accepted 7 August 2007 concerning the higher order classification of basidiomycetes down to order. This paper Published online 16 August 2007 presents a phylogenetic classification for corticioid fungi at the family level. Fifty putative Corresponding Editor: families were identified from published phylogenies and preliminary analyses of unpub- Scott LaGreca lished sequence data. A dataset with 178 terminal taxa was compiled and subjected to phy- logenetic analyses using MP and Bayesian inference. From the analyses, 41 strongly Keywords: supported and three unsupported clades were identified. These clades are treated as fam- Agaricomycetes ilies in a Linnean hierarchical classification and each family is briefly described. Three ad- Basidiomycota ditional families not covered by the phylogenetic analyses are also included in the Molecular systematics classification. All accepted corticioid genera are either referred to one of the families or Phylogeny listed as incertae sedis. Taxonomy ª 2007 The British Mycological Society. Published by Elsevier Ltd. All rights reserved. Introduction develop a downward-facing basidioma. -
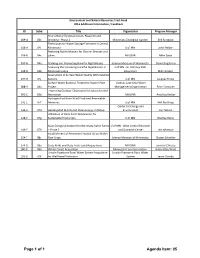
Of 1 Agenda Item: 05 ENRTF ID: 009-A / Subd
Environment and Natural Resources Trust Fund 2016 Additional Information / Feedback ID Subd. Title Organization Program Manager Prairie Butterfly Conservation, Research and 009‐A 03c Breeding ‐ Phase 2 Minnesota Zoological Garden Erik Runquist Techniques for Water Storage Estimates in Central 018‐A 04i Minnesota U of MN John Neiber Restoring Native Mussels for Cleaner Streams and 036‐B 04c Lakes MN DNR Mike Davis 037‐B 04a Tracking and Preventing Harmful Algal Blooms Science Museum of Minnesota Daniel Engstrom Assessing the Increasing Harmful Algal Blooms in U of MN ‐ St. Anthony Falls 038‐B 04b Minnesota Lakes Laboratory Miki Hondzo Assessment of Surface Water Quality With Satellite 047‐B 04j Sensors U of MN Jacques Finlay Surface Water Bacterial Treatment System Pilot Vadnais Lake Area Water 088‐B 04u Project Management Organization Brian Corcoran Improving Outdoor Classrooms for Education and 091‐C 05b Recreation MN DNR Amy Kay Kerber Hydrogen Fuel from Wind Produced Renewable 141‐E 07f Ammonia U of MN Will Northrop Center for Energy and 144‐E 07d Geotargeted Distributed Clean Energy Initiative Environment Carl Nelson Utilization of Dairy Farm Wastewater for 148‐E 07g Sustainable Production U of MN Bradley Heins Solar Energy Utilization for Minnesota Swine Farms U of MN ‐ West Central Research 149‐E 07h – Phase 2 and Outreach Center Lee Johnston Establishment of Permanent Habitat Strips Within 154‐F 08c Row Crops Science Museum of Minnesota Shawn Schottler 174‐G 09a State Parks and State Trails Land Acquisitions MN DNR Jennifer Christie 180‐G 09e Wilder Forest Acquisition Minnesota Food Association Hilary Otey Wold Lincoln Pipestone Rural Water System Acquisition Lincoln Pipestone Rural Water 181‐G 09f for Well Head Protection System Jason Overby Page 1 of 1 Agenda Item: 05 ENRTF ID: 009-A / Subd. -

Conservation Assessment for White Adder's Mouth Orchid (Malaxis B Brachypoda)
Conservation Assessment for White Adder’s Mouth Orchid (Malaxis B Brachypoda) (A. Gray) Fernald Photo: Kenneth J. Sytsma USDA Forest Service, Eastern Region April 2003 Jan Schultz 2727 N Lincoln Road Escanaba, MI 49829 906-786-4062 This Conservation Assessment was prepared to compile the published and unpublished information on Malaxis brachypoda (A. Gray) Fernald. This is an administrative study only and does not represent a management decision or direction by the U.S. Forest Service. Though the best scientific information available was gathered and reported in preparation for this document and subsequently reviewed by subject experts, it is expected that new information will arise. In the spirit of continuous learning and adaptive management, if the reader has information that will assist in conserving the subject taxon, please contact: Eastern Region, USDA Forest Service, Threatened and Endangered Species Program, 310 Wisconsin Avenue, Milwaukee, Wisconsin 53203. Conservation Assessment for White Adder’s Mouth Orchid (Malaxis Brachypoda) (A. Gray) Fernald 2 TABLE OF CONTENTS TABLE OF CONTENTS .................................................................................................................1 ACKNOWLEDGEMENTS..............................................................................................................2 EXECUTIVE SUMMARY ..............................................................................................................3 INTRODUCTION/OBJECTIVES ...................................................................................................3 -

Aegis Boa (Polyporales, Basidiomycota) a New Neotropical Genus and Species Based on Morphological Data and Phylogenetic Evidences
Mycosphere 8(6): 1261–1269 (2017) www.mycosphere.org ISSN 2077 7019 Article Doi 10.5943/mycosphere/8/6/11 Copyright © Guizhou Academy of Agricultural Sciences Aegis boa (Polyporales, Basidiomycota) a new neotropical genus and species based on morphological data and phylogenetic evidences Gómez-Montoya N1, Rajchenberg M2 and Robledo GL 1, 3* 1 Laboratorio de Micología, Instituto Multidisciplinario de Biología Vegetal, Consejo Nacional de Investigaciones Científicas y Técnicas, Universidad Nacional de Córdoba, CC 495, CP 5000 Córdoba, Argentina 2 Centro de Investigación y Extensión Forestal Andino Patagónico (CIEFAP), C.C. 14, 9200 Esquel, Chubut and Universidad Nacional de la Patagonia S.J. Bosco, Ingeniería Forestal, Ruta 259 km 14.6, Esquel, Chubut, Argentina 3 Fundación FungiCosmos, Av. General Paz 54, 4to piso, of. 4, CP 5000, Córdoba, Argentina. Gómez-Montoya N, Rajchenberg M, Robledo GL. 2017 – Aegis boa (Polyporales, Basidiomycota) a new neotropical genus and species based on morphological data and phylogenetic evidences. Mycosphere 8(6), 1261–1269, Doi 10.5943/mycosphere/8/6/11 Abstract The new genus, Aegis Gómez-Montoya, Rajchenb. & Robledo, is described to accommodate the new species Aegis boa based on morphological data and phylogenetic evidences (ITS – LSU rDNA). It is characterized by a particular monomitic hyphal system with thick-walled, widening, inflated and constricted generative hyphae, and allantoid basidiospores. Phylogenetically Aegis is closely related to Antrodiella aurantilaeta, both species presenting an isolated position within Polyporales into Grifola clade. The new taxon is so far known from Yungas Mountain Rainforests of NW Argentina. Key words – Grifola – Neotropical polypores – Tyromyces Introduction Polypore diversity of NW Argentina was reviewed by Robledo & Rajchenberg (2007).