Age-Depended Selection of Chloroquine-Sensitive Plasmodium
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ghana Gazette
GHANA GAZETTE Published by Authority CONTENTS PAGE Facility with Long Term Licence … … … … … … … … … … … … 1236 Facility with Provisional Licence … … … … … … … … … … … … 201 Page | 1 HEALTH FACILITIES WITH LONG TERM LICENCE AS AT 12/01/2021 (ACCORDING TO THE HEALTH INSTITUTIONS AND FACILITIES ACT 829, 2011) TYPE OF PRACTITIONER DATE OF DATE NO NAME OF FACILITY TYPE OF FACILITY LICENCE REGION TOWN DISTRICT IN-CHARGE ISSUE EXPIRY DR. THOMAS PRIMUS 1 A1 HOSPITAL PRIMARY HOSPITAL LONG TERM ASHANTI KUMASI KUMASI METROPOLITAN KPADENOU 19 June 2019 18 June 2022 PROF. JOSEPH WOAHEN 2 ACADEMY CLINIC LIMITED CLINIC LONG TERM ASHANTI ASOKORE MAMPONG KUMASI METROPOLITAN ACHEAMPONG 05 October 2018 04 October 2021 MADAM PAULINA 3 ADAB SAB MATERNITY HOME MATERNITY HOME LONG TERM ASHANTI BOHYEN KUMASI METRO NTOW SAKYIBEA 04 April 2018 03 April 2021 DR. BEN BLAY OFOSU- 4 ADIEBEBA HOSPITAL LIMITED PRIMARY HOSPITAL LONG-TERM ASHANTI ADIEBEBA KUMASI METROPOLITAN BARKO 07 August 2019 06 August 2022 5 ADOM MMROSO MATERNITY HOME HEALTH CENTRE LONG TERM ASHANTI BROFOYEDU-KENYASI KWABRE MR. FELIX ATANGA 23 August 2018 22 August 2021 DR. EMMANUEL 6 AFARI COMMUNITY HOSPITAL LIMITED PRIMARY HOSPITAL LONG TERM ASHANTI AFARI ATWIMA NWABIAGYA MENSAH OSEI 04 January 2019 03 January 2022 AFRICAN DIASPORA CLINIC & MATERNITY MADAM PATRICIA 7 HOME HEALTH CENTRE LONG TERM ASHANTI ABIREM NEWTOWN KWABRE DISTRICT IJEOMA OGU 08 March 2019 07 March 2022 DR. JAMES K. BARNIE- 8 AGA HEALTH FOUNDATION PRIMARY HOSPITAL LONG TERM ASHANTI OBUASI OBUASI MUNICIPAL ASENSO 30 July 2018 29 July 2021 DR. JOSEPH YAW 9 AGAPE MEDICAL CENTRE PRIMARY HOSPITAL LONG TERM ASHANTI EJISU EJISU JUABEN MUNICIPAL MANU 15 March 2019 14 March 2022 10 AHMADIYYA MUSLIM MISSION -ASOKORE PRIMARY HOSPITAL LONG TERM ASHANTI ASOKORE KUMASI METROPOLITAN 30 July 2018 29 July 2021 AHMADIYYA MUSLIM MISSION HOSPITAL- DR. -

'Accelerating Case Detection of Tuberculosis in Ghana' Project
‘Accelerating case detection of tuberculosis in Ghana’ Project Progress Report no. 15 September 2017 27 systems installed ….. In July and early August, 2 fixed systems were installed in Western region (Prestea hospital and Ahmadiyya hospital, Daboase) and 3 containerized systems were installed in Tamale Central hospital (Northern region) and Twifo Praso and Abura Dunkwa in Central region. The installation of a fixed system in Half Assini has been postponed because the hospital could not yet complete their X-ray room. For more information see the overview on page 2. Ahmadiyya hospital Daboase Twifo Praso hospital room installation container installation We are expecting shipment 8 with 4 containerized and 3 fixed systems to arrive around 19th of September and hope to install these systems in October. TB-Screening vans for National Tuberculosis Control Program Early September 2 TB-screening vans arrived at Temporal Central Medical Stores of Ghana Health Service at Spintex road. Engineers from Oldelft and Universal Delft will assemble the X-ray equipment in the vans before the end of the month September. Page 1 of 2 Capacity Building: training for radiographers On 19th and 20th July 15 radiographers from beneficiary hospitals were trained at Universal Delft’s Training Centre in North-Dzorwulu. Main subject was in-depth application training. Other topics addressed, were radiation safety, infection control, CAD4TB and project-information. There was enough time for on-hands training as there is demonstration equipment available at the venue. -

Use of Proscribed Chloroquine Is Associated with an Increased Risk Of
Asare et al. Malaria Journal 2014, 13:246 http://www.malariajournal.com/content/13/1/246 RESEARCH Open Access Use of proscribed chloroquine is associated with an increased risk of pfcrt T76 mutation in some parts of Ghana Kwame K Asare1, Johnson N Boampong1*, Richmond Afoakwah1, Elvis O Ameyaw1, Rakesh Sehgal2 and Neils B Quashie3 Abstract Background: After years of disuse of chloroquine (CQ) as first-line anti-malarial drug in Ghana, reports from molecular studies conducted in parts of the country indicate varying prevalence of T76 mutation in the pfcrt gene. This situation has several health implications, one being that mutations that confer resistance to CQ have been reported to show substantial cross-resistance to other anti-malarial drugs. It is important to identify some of the factors contributing to the continuous presence of CQ resistance markers in the country. This study determined the prevalence of T76 mutation in pfcrt gene of Plasmodium falciparum isolates collected from selected areas of the Central region of Ghana and correlated with the level of CQ use in these areas. Methods: Plasmodium falciparum DNA was extracted from collected blood-blot filter paper samples in the study sites. The prevalence of T76 point mutation in pfcrt gene was assessed using nested PCR followed by RFLP. CQ from pharmacy and chemical shops was obtained using mystery buying method. The extent of CQ use by the participants was determined by measuring the level of the drug in their urine samples using the Saker-Solomon method. Results: Of the 214 P. falciparum isolates analysed, 71.9% were found to have T76 mutation of pfcrt gene. -
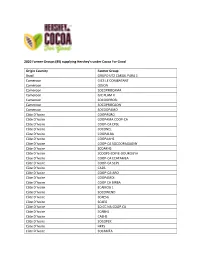
Hershey Cocoa for Good Farmer Groups
2020 Farmer Groups (89) supplying Hershey’s under Cocoa For Good Origin Country Farmer Group Brazil GRUPO UTZ CARGIL PARA 1 Cameroon GICS LE COMBATANT Cameroon ODJON Cameroon SOCOPROCAMA Cameroon GIC PLAM II Cameroon SOCOOPROBI Cameroon SOCOPROCAON Cameroon SOCOOPAMO Côte D’Ivoire COOPAGRO Côte D’Ivoire COOPAMA COOP-CA Côte D’Ivoire COOP-CA CPSL Côte D’Ivoire SOCONEL Côte D’Ivoire COOPALBA Côte D’Ivoire COOPAAHS Côte D’Ivoire COOP-CA SOCOOPAGAGNY Côte D’Ivoire SCOAKHS Côte D’Ivoire SCOOPS-EDIFIE-DOUKOUYA Côte D’Ivoire COOP-CA ECATAMEA Côte D’Ivoire COOP-CA SCPS Côte D’Ivoire CA2B Côte D’Ivoire COOP-CA-ABO Côte D’Ivoire COOPAMDI Côte D’Ivoire COOP CA BIRBA Côte D’Ivoire ECAMOG 1 Côte D’Ivoire SOCOWEND Côte D’Ivoire SCAESG Côte D’Ivoire SCAEG Côte D’Ivoire SO.CC.NA.COOP.CA Côte D’Ivoire SCABHS Côte D’Ivoire CAEHS Côte D’Ivoire SOCOPEK Côte D’Ivoire HKF5 Côte D’Ivoire SOCAMEA Côte D’Ivoire SOCOOPAWA Côte D’Ivoire SOCAHI Côte D’Ivoire CPACD Côte D’Ivoire COOPABKO Côte D’Ivoire SOCOOPADO Côte D’Ivoire ECAL Côte D’Ivoire CAKB Côte D’Ivoire CAPROD Côte D’Ivoire SOCOOPASAHG Côte D’Ivoire CABING Côte D’Ivoire CATEZ Côte D’Ivoire SOCOPROCY Côte D’Ivoire CPAY Côte D’Ivoire COABOB Côte D’Ivoire COOPAAF Côte D’Ivoire CASMA Côte D’Ivoire CANS Côte D’Ivoire SCAC Côte D’Ivoire COOP-CA NANTEBESSOU Côte D’Ivoire SOCOPADOU Côte D’Ivoire SOCOOPELA Côte D’Ivoire SOCAT Côte D’Ivoire SOCOEXPAHS Côte D’Ivoire CABA Côte D’Ivoire CAREFSI Côte D’Ivoire COOPATESA Côte D’Ivoire CAABRO Côte D’Ivoire CAEVA Côte D’Ivoire SCOOPS JPK Côte D’Ivoire COOPAKAM Côte D’Ivoire COOPAOU Côte D’Ivoire COALIDAN Côte D’Ivoire COOP TEEGA WENDE Côte D’Ivoire SCAAO Côte D’Ivoire SCOOPADIM Côte D’Ivoire SCOOPS CARET Ecuador OLAM ECUADOR S.A. -

Download PDF (367.6
©> f GAZETTi - •' r J- Published by Authority i No. 31 FRIDAY, 4th AUGUST 20 uO CONTENTS Page Notice 61'Puhlicaiion of an Executive Instrument.. 268 Notice of Publication of a Bill 268 The National Weekly Lotto Results 268 Licence lor the Celebration of Marriages—Public Place of Worship lRef Holy Ghost Chapel International, Asawasi, Kuniasi) 268 Licence for the Celebration of Marriages—Public Place of Worship (Ref. International Life Gate Chapel, La) 268 Change of Names 268 V . GHANA GAZETTE, 4th AUGUST, 2000_______ r 268 NOTICE OF PUBLICATION OF EXECUTIVE INSTRUMENT The following Executive Instrument is published today: Deportation Order, 2000 (E.I. 14) NOTICE OF PUBLICATION OF A BILL The following Bill is published today: The Interpretation Bill THE NATIONAL WEEKLY LOTTO RESULTS It is hereby certified that a National Weekly Lotto Draw was held on Saturday, 29th July 2000 in accordance with the provisions of the National Weekly Lotto Act, 1961 and the Regulations published thereunder and that the following numbers were drawn in successive order: 82-9-90-60-5 Col. MARTIN GBIKPI Director of National Lotteries PUBLIC PLACE OF WORSHIP LICENCE FOR THE CELEBRATION OF MARRIAGES Under the provision of section 8, subsection (1) of the Marriages Ordinance (Cap. 127) as amended. I, Gershon Koku Kumor, Assistant Director I. Office of the Ashanti Regional Co-ordinating Council, do hereby license the following public place of worship for the celebration of Marriages: Holy Ghost Chapel International Asawasi P.O. Box 3823 Kumasi. Given under my hand at the office of the Ashanti Regional Co-ordinating Council, this 24th day of March, 2000. -

National Flood Hazard
3°0'0"W 2°0'0"W 1°0'0"W 0°0'0" 1°0'0"E National Flood Hazard Map - Current Situation (2010) ´ Burkina Faso Pusiga Bawku N N " " 0 0 ' ' 0 0 ° Gwollu Paga ° 1 1 1 1 Zebilla Bongo Navrongo Tumu Nangodi Nandom Garu Lambusie Bolgatanga Sandema ^_ Tongo Lawra Jirapa Gambaga Bunkpurugu Fumbisi Issa Nadawli Walewale Funsi Yagaba Chereponi ^_Wa N N " " 0 0 ' ' 0 0 ° ° 0 0 1 1 Karaga Gushiegu Wenchiau Saboba Savelugu Kumbungu Daboya Yendi Tolon Sagnerigu Tamale Sang ^_ Tatale Zabzugu Sawla Damongo Bole N N " " 0 0 ' ' 0 0 ° ° 9 9 Bimbila Buipe Wulensi Togo Salaga Kpasa Kpandai Côte d'Ivoire Nkwanta Yeji Banda Ahenkro Chindiri Dambai Kintampo N N " " 0 0 ' ' 0 0 ° ° 8 Sampa 8 Jema Nsawkaw Kete-krachi Kajeji Atebubu Wenchi Kwame Danso Busunya Drobo Techiman Nkoranza Kadjebi Berekum Akumadan Jasikan Odumase Ejura Sunyani Wamfie ^_ Dormaa Ahenkro Duayaw Nkwanta Hohoe Bechem Nkonya Ahenkro Mampong Ashanti Drobonso Donkorkrom Nkrankwanta N N " Tepa Nsuta " 0 Va Golokwati 0 ' Kpandu ' 0 0 ° Kenyase No. 1 ° 7 7 Hwediem Ofinso Tease Agona AkrofosoKumawu Anfoega Effiduase Adaborkrom Mankranso Kodie Goaso Mamponteng Agogo Ejisu Kukuom Kumasi Essam- Debiso Nkawie ^_ Abetifi Kpeve Foase Kokoben Konongo-odumase Nyinahin Ho Juaso Mpraeso ^_ Kuntenase Nkawkaw Kpetoe Manso Nkwanta Bibiani Bekwai Adaklu Waya Asiwa Begoro Asesewa Ave Dapka Jacobu New Abirem Juabeso Kwabeng Fomena Atimpoku Bodi Dzodze Sefwi Wiawso Obuasi Ofoase Diaso Kibi Dadieso Akatsi Kade Koforidua Somanya Denu Bator Dugame New Edubiase ^_ Adidome Akontombra Akwatia Suhum N N " " 0 Sogakope 0 -

Presentation (PDF)1.24 MB
Improved Access to Medicines through Local Production Martha Gyansa-Lutterodt Director of Pharmaceutical Services Ministry of Health, Ghana 3/21/2013 Introduction • Access to medicines remain a global challenge; and affect LMICs to varying extents, Ghana inclusive • Local manufacture of medicines contribute significantly to the improvement of access to medicines through simple supply chains • The interphase between private sector and public sector to improve access remain a potential yet to be explored fully • Thus the interlock between access to medicines and the rate of local production; and quality and capacity building are areas to be explored for Ghana 3/21/2013 Introduction (2) • The health challenge in Ghana major factor in poverty • Many deaths preventable with timely access to appropriate & affordable medicines • Ghana depends largely on imports from Asia, frequently with long lead times • Still have challenges with quality of some imports 3/21/2013 Ghana Map #BAWKU #BONGO #ZEBILLA #TUMU #NAVRONGO # #SANDEMA B#YOLGATANGA #LAWRA LAWRA # Upper East #GAMBAGA NADAWLI # #WALEWALE Upper West #Y# WA #GUSHIEGU #SABOBA Northern #SAVELUGU TOLON TAMALE YENDI # #Y# # #ZABZUGU #DAMONGO #BOLE #BIMBILA #SALAGA #NKWANTA #KINTAMPO Brong Ahafo #KETE-KRACHI #WENCHI #ATEBUBU #KWAME DANSO TECHIMAN #DROBO # #NKORANZA #KADJEBI # BEREKUM # #EJURA JASIKAN #SUNYANI #DORMAA AHENKRO #Y #HOHOE #BECHEM Ashanti DONKORKROM # #TEPA # #KENYASE NO. 1 OFINSO KPANDU # #AGONA AKROFOSO MANK#RANSO #EFFIDUASE #GOASO #MAMPONTENG KUM#ASI#EJISU # #Y KONONGO-ODUMASE #MAMPONG -

Staff-ID-Central-Region
CENTRAL-882 S/N STAFF ID FULL_NAME MANAGEMENT_UNIT DISTRICT 1 1419487 MENSAH, Mr. ERIC ABONIN 0307 Abirem-Agona Meth JHS (METH) Komenda-Edina-Eguafo-Abirem Municipal Assembly 2 1418761 ASIEDU, Miss GIFTY 0307 Abrem Essiam DC JHS (GOV) Komenda-Edina-Eguafo-Abirem Municipal Assembly 3 1419605 ADDAE, Mr. ENOCK 0307 Abrem-Agona Cath Prim (CATH) Komenda-Edina-Eguafo-Abirem Municipal Assembly 4 1419080 DONKOR, Miss MABEL BOAKYE 0307 Abrem-Agona Meth Prim (METH) Komenda-Edina-Eguafo-Abirem Municipal Assembly 5 1419053 FORSON, Mr. FRANCIS 0307 Abrobiano MA Prim (GOV) Komenda-Edina-Eguafo-Abirem Municipal Assembly 6 1419451 AMPOMAH, Miss ANITA 0307 Abrobiano MA Prim (GOV) Komenda-Edina-Eguafo-Abirem Municipal Assembly 7 1419519 AMARH-AKU, Miss SANDRA AMARTELEY 0307 Abrobiano MA Prim (GOV) Komenda-Edina-Eguafo-Abirem Municipal Assembly 8 1418712 OTCHERE, Mr. DOMINIC OTABIL 0307 Abrobiano Isl Prim (ISL) Komenda-Edina-Eguafo-Abirem Municipal Assembly 9 1419540 TECHIE-MENSAH, Mr. WAATEMBER 0307 Abrobiano Isl Prim (ISL) Komenda-Edina-Eguafo-Abirem Municipal Assembly 10 1419588 OMANE ACHEAMFOUR, Miss ERNESTINA 0307 Abrobiano Isl Prim (ISL) Komenda-Edina-Eguafo-Abirem Municipal Assembly 11 1417781 ARTHUR, Miss GIFTY 0306 Ajumako Ansa-Bre DA JHS (GOV) Gomoa West 12 1418899 TWUMASI, Mr. CLEMENT 0307 Ampenyi DC JHS (GOV) Komenda-Edina-Eguafo-Abirem Municipal Assembly 13 1418782 AGYEIWAA, Miss SANDRA 0307 Ampenyi DC JHS (GOV) Komenda-Edina-Eguafo-Abirem Municipal Assembly 14 1418981 HOWARD, Miss ANITA 0307 Anweem-Kissi MA Prim (GOV) Komenda-Edina-Eguafo-Abirem Municipal Assembly 15 1417599 BAIDOO, Miss FAUSTINA 0306 Asempanyin DA Prim (GOV) Gomoa West 16 1417577 AMPONSAH, Miss NANCY 0306 Asempanyin DA Prim (GOV) Gomoa West 17 1417609 FOSU, Mr. -

Teacher Licensing Centres and Locations Central Region
TEACHER LICENSING CENTRES AND LOCATIONS CENTRAL REGION TEACHER LICENSING CENTRES AND LOCATIONS (JUNE 2021) CENTRAL REGION DATES MMDs CENTRE NAMES DIRECTIONS 01/06 & AWUTU ST. MARTHA'S CATHOLIC BASIC ON OBOM ROAD BEFORE 02/06 SENYA BAWJIASE JN, THEN TURN EAST RIGHT FROM RENT CONTROL BUILDING 01/06 & AWUTU ODUPONGKPEHE M/A BASIC ON NYANYANO ROAD NEAR 02/06 SENYA SCHOOL CANTEEN THE FIRST TRAFFIC LIGHT EAST 01/06 & AWUTU ST.MARY'S ANGLICAN JHS KASOA BAWJIASE ROAD 02/06 SENYA EAST DATES MMDs CENTRE NAMES DIRECTIONS 01/06 & AWUTU ODUPONG SHS FROM St, MARY'S ANGLICAN 02/06 SENYA CONTINUE TO OFAAKOR & EAST BRANCH LEFT 01/06 & AWUTU BAWJIASE METHODIST BASIC BAWJIASE TOWNSHIP 02/06 SENYA SCH WEST 01/06 & AWUTU BAWJIASE D/A BASIC SCH BAWJIASE TOWNSHIP 02/06 SENYA WEST 01/06 & AWUTU DISTRICT EDUCATION ON AWUTU- 02/06 SENYA CONF.HALL BEREKU/BONTRASE RD WEST CLOSE TO THE COURT 01/06 & AWUTU OBRAKYIRE D/A SCH ON THE AWUTU/BREKU- 02/06 SENYA BONTRASE ROAD WEST 01/06 & GOMOA EAST THE HARTLEY TRUST BRANCH LEFT AT KASOA 02/06 FOUNDATION SCH TO NYANYANO 01/06 & GOMOA EAST ST. GREGORY CATH. BUDUBURAM ON THE 02/06 CHURCH KASOA WINNEBA ROAD 01/06 & GOMOA EAST OJOBI D/A BASIC AT AKOTSI JN. AFTER 02/06 SCH BUDUBURAM ASK OF DIRECTION TO OJOBI 01/06 & GOMOA EAST POTSIN METHODIST AT GOMOA POTSIN ON 02/06 CHURCH THEKASOA WINNEBA ROAD DATES MMDs CENTRE NAMES DIRECTIONS 03/06 & AGONA WEST SWEDRU EMMANUEL NEAR SWEDRU ESG 04/06 METHODIST CHURCH OFFICE (POWER HOUSE) 03/06 & AGONA WEST SWEDRU SDA CLOSE TO SWEDRU PIPE 04/06 CHURCH TANK 03/06 & AGONA WEST NYAKROM NEAR NYAKROM 04/06 METHODIST CHURCH VICTORIA PARK 03/06 & AGONA WEST FANKOBAA SHS FROM NYAKROM, TURN 04/06 LEFT TO BOBIKUMA, THEN LEFT TO FANKOBAA SHS 03/06 & AGONA EAST NSABA PRESBY AGONA NSABA 04/06 CHURCH 03/06 & AGONA EAST DUAKWA PRESBY AGONA DUAKWA 04/06 CHURCH 03/06 & AGONA EAST NAMANWORA COMM. -

Mapping Forest Landscape Restoration Opportunities in Ghana
MAPPING FOREST LANDSCAPE RESTORATION OPPORTUNITIES IN GHANA 1 Assessment of Forest Landscape Restoration Assessing and Capitalizing on the Potential to Potential In Ghana To Contribute To REDD+ Enhance Forest Carbon Sinks through Forest Strategies For Climate Change Mitigation, Landscape Restoration while Benefitting Poverty Alleviation And Sustainable Forest Biodiversity Management FLR Opportunities/Potential in Ghana 2 PROCESS National Assessment of Off-Reserve Areas Framework Method Regional Workshops National National National - Moist Stakeholders’ Assessment of validation - Transition Workshop Forest Reserves Workshop - Savannah - Volta NREG, FIP, FCPF, etc 3 INCEPTION WORKSHOP . Participants informed about the project . Institutional commitments to collaborate with the project secured . The concept of forest landscape restoration communicated and understood . Forest condition scoring proposed for reserves within and outside the high forest zone 4 National Assessment of Forest Reserves 5 RESERVES AND NATIONAL PARKS IN GHANA Burkina Faso &V BAWKU ZEBILLA BONGO NAVRONGO TUMU &V &V &V &V SANDEMA &V BOLGATANGA &V LAWRA &V JIRAPA GAMBAGA &V &V N NADAWLI WALEWALE &V &V WA &V GUSHIEGU &V SABOBA &V SAVELUGU &V TOLON YENDI TAMALE &V &V &V ZABZUGU &V DAMONGO BOLE &V &V BIMBILA &V Republic of SALAGA Togo &V NKWANTA Republic &V of Cote D'ivoire KINTAMPO &V KETE-KRACHI ATEBUBU WENCHI KWAME DANSO &V &V &V &V DROBO TECHIMAN NKORANZA &V &V &V KADJEBI &V BEREKUM JASIKAN &V EJURA &V SUNYANI &V DORMAA AHENKRO &V &V HOHOE BECHEM &V &V DONKORKROM TEPA -

Assessment of Forest Landscape Restoration Opportunities in Ghana
ASSESSMENT OF FOREST LANDSCAPE RESTORATION OPPORTUNITIES IN GHANA Final Report SEPTEMBER 2011 List of Acronyms CBO Community Based Organization CERSGIS Centre for Remote Sensing and Geographic Information Services CREMA Community Resource Management Areas DBH Diameter at Breast Height/Base Diameter EI Economic Index EU European Union FC Forestry Commission FLEGT Forest Law Enforcement, Governance and Trade FORIG Forestry Research Institute of Ghana FSD Forest Services Division GHI Genetic Heat Index GIZ Deutsche Gesellschaft für Internationale Zusammenarbeit HFZ High Forest Zone ITTO International Tropical Timber Organization IUCN International Union for Conservation of Nature MTS Modified Taungya System NCRC Nature Conservation Research Centre NGO Non Governmental Organization N-REG Natural Resources and Environmental Governance PI Pioneer Index PROFOR Program on Forests REDD Reducing Emissions from Deforestation and Forest Degradation RMSC Resource Management Support Center of the Forestry Commission SDSU South Dakota State University TZ Transition Zone UNFCCC United Nations Framework Convention on Climate Change UNESCO United Nations Educational, Scientific and Cultural Organization WD Wildlife Division of the Forestry Commission WRI World Resources Institute 2 Table of Contents List of Acronyms ............................................................................................................................. 2 Table of Contents ........................................................................................................................... -

Ministry of Health
REPUBLIC OF GHANA MEDIUM TERM EXPENDITURE FRAMEWORK (MTEF) FOR 2021-2024 MINISTRY OF HEALTH PROGRAMME BASED BUDGET ESTIMATES For 2021 Transforming Ghana Beyond Aid REPUBLIC OF GHANA Finance Drive, Ministries-Accra Digital Address: GA - 144-2024 MB40, Accra - Ghana +233 302-747-197 [email protected] mofep.gov.gh Stay Safe: Protect yourself and others © 2021. All rights reserved. No part of this publication may be stored in a retrieval system or Observe the COVID-19 Health and Safety Protocols transmitted in any or by any means, electronic, mechanical, photocopying, recording or otherwise without the prior written permission of the Ministry of Finance Get Vaccinated MINISTRY OF HEALTH 2021 BUDGET ESTIMATES The MoH MTEF PBB for 2021 is also available on the internet at: www.mofep.gov.gh ii | 2021 BUDGET ESTIMATES Contents PART A: STRATEGIC OVERVIEW OF THE MINISTRY OF HEALTH ................................ 2 1. NATIONAL MEDIUM TERM POLICY OBJECTIVES ..................................................... 2 2. GOAL ............................................................................................................................ 2 3. VISION .......................................................................................................................... 2 4. MISSION........................................................................................................................ 2 5. CORE FUNCTIONS ........................................................................................................ 2 6. POLICY OUTCOME