Supplementary Materials and Methods
Plasmids. The table below contains information about all the plasmids utilized in our studies.
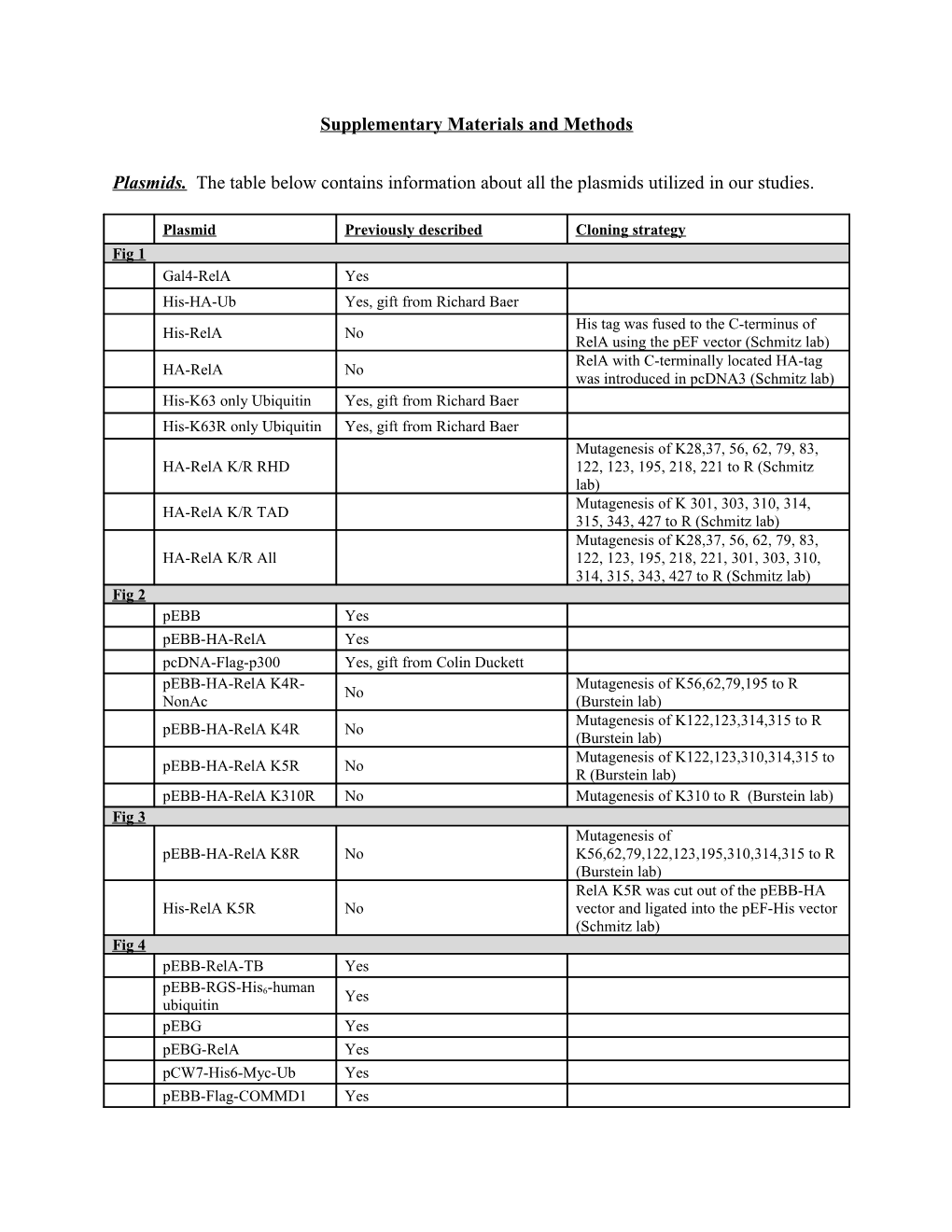
Plasmid Previously described Cloning strategy Fig 1 Gal4-RelA Yes His-HA-Ub Yes, gift from Richard Baer His tag was fused to the C-terminus of His-RelA No RelA using the pEF vector (Schmitz lab) RelA with C-terminally located HA-tag HA-RelA No was introduced in pcDNA3 (Schmitz lab) His-K63 only Ubiquitin Yes, gift from Richard Baer His-K63R only Ubiquitin Yes, gift from Richard Baer Mutagenesis of K28,37, 56, 62, 79, 83, HA-RelA K/R RHD 122, 123, 195, 218, 221 to R (Schmitz lab) Mutagenesis of K 301, 303, 310, 314, HA-RelA K/R TAD 315, 343, 427 to R (Schmitz lab) Mutagenesis of K28,37, 56, 62, 79, 83, HA-RelA K/R All 122, 123, 195, 218, 221, 301, 303, 310, 314, 315, 343, 427 to R (Schmitz lab) Fig 2 pEBB Yes pEBB-HA-RelA Yes pcDNA-Flag-p300 Yes, gift from Colin Duckett pEBB-HA-RelA K4R- Mutagenesis of K56,62,79,195 to R No NonAc (Burstein lab) Mutagenesis of K122,123,314,315 to R pEBB-HA-RelA K4R No (Burstein lab) Mutagenesis of K122,123,310,314,315 to pEBB-HA-RelA K5R No R (Burstein lab) pEBB-HA-RelA K310R No Mutagenesis of K310 to R (Burstein lab) Fig 3 Mutagenesis of pEBB-HA-RelA K8R No K56,62,79,122,123,195,310,314,315 to R (Burstein lab) RelA K5R was cut out of the pEBB-HA His-RelA K5R No vector and ligated into the pEF-His vector (Schmitz lab) Fig 4 pEBB-RelA-TB Yes pEBB-RGS-His -human 6 Yes ubiquitin pEBG Yes pEBG-RelA Yes pCW7-His6-Myc-Ub Yes pEBB-Flag-COMMD1 Yes pEBB-Flag-Cul2 Yes pEBB-Flag GCN5 E575Q Yes YFP-CBP Yes Fig 5 FG9 Yes PCR from Image clone 3489295 (Burstein FG9-HA-mRelA WT No lab) FG9-HA-mRelA K310R No Mutagenesis of K310 to R (Burstein lab) Mutagenesis of K122,123,314,315 to R FG9-HA-mRelA K4R No (Burstein lab) Mutagenesis of K122,123,310,314,315 to FG9-HA-mRelA K5R No R (Burstein lab) FG9-HA-mRelA K310Q No Mutagenesis of K310 to Q (Burstein lab) Mutagenesis of K122,123,314,315 to Q FG9-HA-mRelA K4Q No (Burstein lab) Mutagenesis of K122,123,310,314,315 to FG9-HA-mRelA K5Q No Q (Burstein lab) Antibodies. The following antibodies were utilized for immunoblotting in our studies.
Antibody Supplier Cat. No. Fig 1 Ubiquitin (P4D1) Cell Signaling 3936 RelA (C-Term, C-20) Santa Cruz Biotechnology sc-372 RelA (N-Term, F-6) Santa Cruz Biotechnology sc-8008 Tubulin Sigma T4026 K63-only Ubiquitin Biomol PW0600 Fig 2 Acetylated lysine Cell Signaling 9441 FLAG Sigma A8592 HA CoVance MMS101R Fig 3 Ubiquitin (P4D1) Cell Signaling 3936 HA Covance MMS101R RelA (C-Term, C-20) Santa Cruz Biotechnology sc-372 K63-only Ubiquitin Biomol PW0600 Tubulin Sigma T4026 Fig 4 Streptavidin-HRP BioLegend 405210 Acetylated lysine Cell Signaling 9441 FLAG Sigma A8592 GST Santa Cruz Biotechnology sc-459 HA Covance MMS101R Ubiquitin (P4D1) Cell Signaling 3936 GFP Roche 11814460001.00 Fig 5 HA Covance MMS101R RelA Santa Cruz Biotechnology sc-8008 IB- Upstate Biotechnologies 06-494 IB- Santa Cruz Biotechnology sc-9130 IB- Santa Cruz Biotechnology sc-7155 p50 Santa Cruz Biotechnology sc-8414 p52 Santa Cruz Biotechnology sc-848 RelB Santa Cruz Biotechnology sc-226 c-Rel Santa Cruz Biotechnology sc-6955 -Actin Sigma A5441 Fig S1 RelA(C-Term, C-20) Santa Cruz Biotechnology sc-372 Tubulin Sigma T4026 HDAC1 (H11) Santa Cruz G1008 Primers. The following table provides information regarding the qRT-PCR and ChIP primers utilized in our studies. qRT-PCR Gene Sense primer Antisense primer Reference target (mouse) Nfkbia CGCAGACCTGCACACCCCAG GGAGGGCTGTCCGGCCATTG This study Vcam1 AGTTGGGGATTCGGTTGTTCT CCCCTCATTCCTTACCACCC (Moreno et al 2010) Icam1 GGAGACGCAGAGGACCTTAAC CGCTCAGAAGAACCACCTTC (Geng et al 2009) Mmp13 ACCTCCACAGTTGACAGGCT AGGCACTCCACATCTTGGTTT (Moreno et al 2010) Saa3 CTGTTCAGAAGTTCACGGGAC AGCAGGTCGGAAGTGGTT (Moreno et al 2010) Il6 TGGATGCTACCAAACTGGAT GGACTCTGGCTTTGTCTTTC (Moreno et al 2010) Lamb3 GGCTGCCTCGAAATTACAACA ACCCTCCATGTCTTGCCAAAG Primerbank ID: 6678660a1 Cxcl10 AGTGAACTGCGCTGTCAATG CTTCAGGGTCAAGGCAAACT (Moreno et al 2010) Mmp3 ACCTATTCCTGGTTGCTG GCCTTGGCTGAGTGGTAG (Moreno et al 2010) Mmp9 CGAACTTCGACACTGACAAGAAGT GCACGCTGGAATGATCTAAGC Expi GGAGATGGATCGTGCTCTGG GGCTAGCCATCAGTCCTGC Ccl9 CCAGTGGTGGGTGTACCAG CTCCGATCACTGGGGTTG ChIP Gene promoter Sense primer Antisense primer Reference (species) Vcam1 (mouse) TCAGCCCAGAAAGCAGC AAAGTGTTCAGCCTCCA (Moreno et al 2010) Icam1 (mouse) AGGGGACTAGGCAGTAGTCAATCAG GAACGAGGGCTTCGGTATTT (Geng et al 2009) ICAM1 (human) CCCGATTGCTTTAGCTTGGAA CCGGAACAAATGCTGCAGTTAT (Horion et al 2007) Mmp13 (mouse) GCAGACATTTTCCTTA ATGTTTGTGACTTGGA (Moreno et al 2010) Microarray analysis
RNA extraction and quality control. RNA was extracted using the Trizol method according to the manufacturer’s instructions (Invitrogen; Carlsbad, California, USA). RNA integrity was validated using an Agilent Bioanalyzer. All RNA preparations had RIN values above 8.8 except for samples from rela-/- fibroblasts reconstituted with vector control and treated for 8h with TNF.
The latter showed substantial degradation of total RNA as expected from TNF-dependent apoptosis in the absence of protection by NF-B/RelA dependent gene expression. However, these samples could also be subjected to the microarray analysis protocol.
Microarray-based mRNA expression analysis (Single Color Mode). The “Whole Mouse
Genome Oligo Microarray V2” (G4846A, ID 026655, Agilent Technologies; Santa Clara,
California, USA) used in this study contains 44397 oligonucleotide probes covering the entire murine transcriptome. Synthesis of Cy3-labeled cRNA was performed with the “Quick Amp
Labeling kit, one color” (#5190-0442, Agilent Technologies) according to the manufacturer’s recommendations. cRNA fragmentation, hybridization and washing steps were also carried-out exactly as recommended: “One-Color Microarray-Based Gene Expression Analysis Protocol
V5.7” (see http://www.agilent.com for details). Slides were scanned on the Agilent Micro Array
Scanner G2565CA (pixel resolution 5 µm, bit depth 20). Data extraction was performed with the
“Feature Extraction Software V10.7.3.1” by using the recommended default extraction protocol file: GE1_107_Sep09.xml.
Normalization. The single channel data generated by the Feature Extraction software were normalized and analyzed in Genespring GX software, version 11.5.1 (Agilent Technologies). Expression values were log2 transformed, normalized to 75th percentile of each array and inter array median centered according to the standard procedure in Genespring GX.
Identification of RelA- and K5-dependent gene sets. According to their annotation in
EntrezGene, out of 44397 probes 35843 probes corresponding to 28299 genes were selected. Of these, 24902 probes corresponding to 19833 genes showed hybridization signals above the 20% percentile in at least 4 of the 24 microarray hybridizations and were marked as measurable fluorescence intensity values in at least 16 out of 24 hybridizations. In this data set 419 probes corresponding to 398 genes were regulated by 1h or 8h TNF stimulation by at least two-fold in both independent experiments in rela-deficient fibroblasts stably reconstituted with wild type mRelA. In addition to the regulation by TNF, these genes showed a mean raw expression value above or equal to 50 fluorescence units across all 24 hybridizations. Thereof a total of 197 probe values corresponding 185 genes indicated differential regulation by two-fold compared to averaged values from wild type-reconstituted cells in at least one of the experiments using K5 mutants at least one time point. For these 185 genes, average ratios values of the wild-type reconstituted cells, and, individual ratio values for either K5 reconstituted cell line compared to the WT-reconstituted cells, were analyzed by hierarchical cluster analysis using MeV
MultiExperimentViewer, version 4.6.2, 2011, (www.tm4.org) with cluster settings “complete” as linkage method and “Pearson correlation” for distance measure . A distance threshold of 2.065 resulted in 10 gene clusters, marked by blue triangles as shown in Fig. 5E. Ratio values for rela-/- cells were not included in the cluster analysis but are displayed along the gene list to facilitate identification of RelA-dependent genes. Chromatin immunoprecipitation.
Cells were cross-linked with 1% (v/v) formaldehyde, collected and then lysed in RIPA buffer [10 mM Tris pH 7.5, 150 mM NaCl, 1% (v/v) NP-40, 1% (w/v) Na-Deoxycholate, 0.1% (w/v) SDS,
1 mM EDTA and 10 µg/ml aprotinin]. DNA was sheared by sonification using a Branson sonifier 250. After removal of cellular debris by centrifugation, equal amounts of DNA were incubated with 2 μg of anti-RelA, anti-HA, anti-CBP, anti-Acetyl H3 or control IgG antibodies previously bound to ChIP-grade protein G-coupled magnetic beads (Cell Signaling; Danvers,
Massachusetts, USA). After rotating for 2 h at 4°C on a spinning wheel, the magnetic beads were washed five times using the following buffers: RIPA buffer (twice), RIPA high-salt buffer
[2 M NaCl, 10 mM Tris pH 7.5, 1% (v/v) NP-40, 0.5% (v/v) Na-Deoxycholate and 1 mM
EDTA], RIPA and TE buffer (10 mM Tris pH 7.5 and 1 mM EDTA). The precipitated fragments were eluted with TE buffer containing 1% SDS. For re-ChIP assays, the eluted material was diluted 1:10 with RIPA buffer lacking SDS and precipitated again with the anti-
RelA antibody. The eluted DNA was quantified by real-time PCR using specific primer sets flanking the expected RelA binding site. The signal was normalized by the background IgG precipitation and expressed as fold of the WT unstimulated sample. The following antibodies were used for immunoprecipitations: ChIP-grade HA (Abcam ab 9110), ChIP grade RelA (C-20,
Santa Cruz sc-372x), CBP (C-20, Santa Cruz sc-583), acetyl-Histone H3 (Milipore 06-599). References
Burstein E, Ganesh L, Dick RD, van De Sluis B, Wilkinson JC, Klomp LW et al (2004). A novel role for XIAP in copper homeostasis through regulation of MURR1. EMBO Journal 23: 244- 254.
Burstein E, Hoberg JE, Wilkinson AS, Rumble JM, Csomos RA, Komarck CM et al (2005). COMMD proteins: A novel family of structural and functional homologs of MURR1. J Biol Chem 280: 22222-22232.
Geng H, Wittwer T, Dittrich-Breiholz O, Kracht M, Schmitz ML (2009). Phosphorylation of NF- B p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep 10: 381-386.
Horion J, Gloire G, El Mjiyad N, Quivy V, Vermeulen L, Vanden Berghe W et al (2007). Histone deacetylase inhibitor trichostatin A sustains sodium pervanadate-induced NF-B activation by delaying IBmRNA resynthesis: comparison with tumor necrosis factor . J Biol Chem 282: 15383-15393.
Korsten H, Ziel-van der Made A, Ma X, van der Kwast T, Trapman J (2009). Accumulating progenitor cells in the luminal epithelial cell layer are candidate tumor initiating cells in a Pten knockout mouse prostate cancer model. PLoS One 4: e5662.
Maine GN, Mao X, Komarck CM, Burstein E (2007). COMMD1 promotes the ubiquitination of NF-B subunits through a Cullin-containing ubiquitin ligase. EMBO Journal 26: 436-447.
Maine GN, Li H, Zaidi IW, Basrur V, Elenitoba-Johnson KS, Burstein E (2010). A bimolecular affinity purification method under denaturing conditions for rapid isolation of a ubiquitinated protein for mass spectrometry analysis. Nat Protoc 5: 1447-1459.
Mao X, Gluck N, Li D, Maine GN, Li H, Zaidi IW et al (2009). GCN5 is a required cofactor for a ubiquitin ligase that targets NF-B/RelA. Genes Dev 23: 849-861.
Mizushima S, Nagata S (1990). pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res 18: 5322.
Moreno R, Sobotzik JM, Schultz C, Schmitz ML (2010). Specification of the NF-B transcriptional response by p65 phosphorylation and TNF-induced nuclear translocation of IKK. Nucleic Acids Res 38: 6029-6044.
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N et al (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374-378. Sanchez I, Hughes RT, Mayer BJ, Yee K, Woodgett JR, Avruch J et al (1994). Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature 372: 794-798.
Schmitz ML, Baeuerle PA (1991). The p65 subunit is responsible for the strong transcription activating potential of NF-B. EMBO J 10: 3805-3817.
Spandidos A, Wang X, Wang H, Seed B (2010). PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res 38: D792- D799.
Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P (2002). Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell 9: 265- 277. van Erp K, Dach K, Koch I, Heesemann J, Hoffmann R (2006). Role of strain differences on host resistance and the transcriptional response of macrophages to infection with Yersinia enterocolitica. Physiol Genomics 25: 75-84.
Wells JE, Rice TK, Nuttall RK, Edwards DR, Zekki H, Rivest S et al (2003). An adverse role for matrix metalloproteinase 12 after spinal cord injury in mice. J Neurosci 23: 10107-10115.
Wright CW, Duckett CS (2009). The aryl hydrocarbon nuclear translocator alters CD30- mediated NF-B-dependent transcription. Science 323: 251-255.
Yu H, Kopito RR (1999). The role of multiubiquitination in dislocation and degradation of the alpha subunit of the T cell antigen receptor. J Biol Chem 274: 36852-36858.