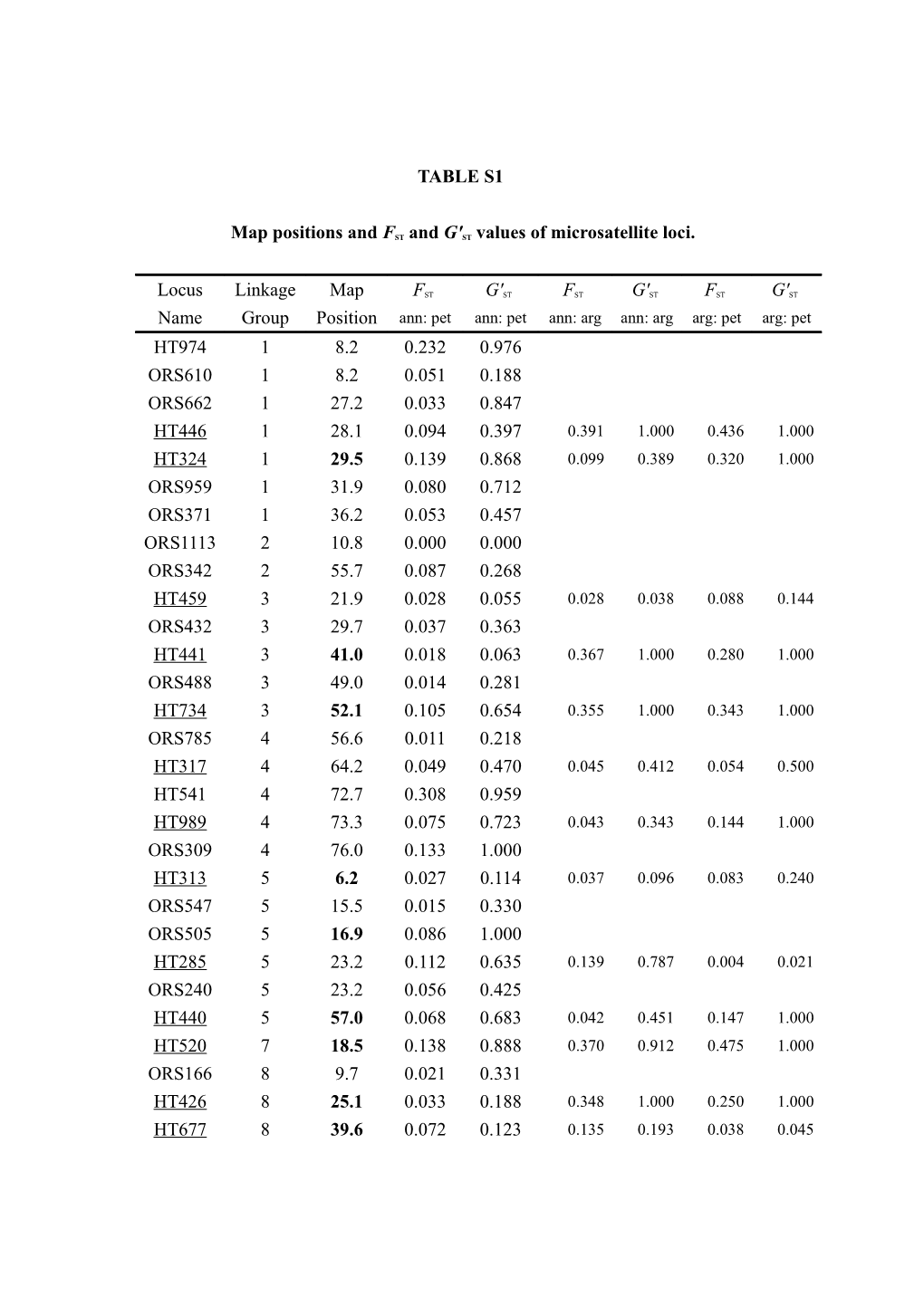
TABLE S1
Map positions and FST and G'ST values of microsatellite loci.
Locus Linkage Map FST G'ST FST G'ST FST G'ST Name Group Position ann: pet ann: pet ann: arg ann: arg arg: pet arg: pet HT974 1 8.2 0.232 0.976 ORS610 1 8.2 0.051 0.188 ORS662 1 27.2 0.033 0.847 HT446 1 28.1 0.094 0.397 0.391 1.000 0.436 1.000 HT324 1 29.5 0.139 0.868 0.099 0.389 0.320 1.000 ORS959 1 31.9 0.080 0.712 ORS371 1 36.2 0.053 0.457 ORS1113 2 10.8 0.000 0.000 ORS342 2 55.7 0.087 0.268 HT459 3 21.9 0.028 0.055 0.028 0.038 0.088 0.144 ORS432 3 29.7 0.037 0.363 HT441 3 41.0 0.018 0.063 0.367 1.000 0.280 1.000 ORS488 3 49.0 0.014 0.281 HT734 3 52.1 0.105 0.654 0.355 1.000 0.343 1.000 ORS785 4 56.6 0.011 0.218 HT317 4 64.2 0.049 0.470 0.045 0.412 0.054 0.500 HT541 4 72.7 0.308 0.959 HT989 4 73.3 0.075 0.723 0.043 0.343 0.144 1.000 ORS309 4 76.0 0.133 1.000 HT313 5 6.2 0.027 0.114 0.037 0.096 0.083 0.240 ORS547 5 15.5 0.015 0.330 ORS505 5 16.9 0.086 1.000 HT285 5 23.2 0.112 0.635 0.139 0.787 0.004 0.021 ORS240 5 23.2 0.056 0.425 HT440 5 57.0 0.068 0.683 0.042 0.451 0.147 1.000 HT520 7 18.5 0.138 0.888 0.370 0.912 0.475 1.000 ORS166 8 9.7 0.021 0.331 HT426 8 25.1 0.033 0.188 0.348 1.000 0.250 1.000 HT677 8 39.6 0.072 0.123 0.135 0.193 0.038 0.045 HT668 8 78.5 0.054 0.965 0.450 1.000 0.449 1.000 HT679 9 37.6 0.021 0.027 HT834 9 42.7 0.143 0.974 0.056 0.396 0.039 0.322 ORS275 9 75.0 0.034 0.086 HT978 9 84.9 0.137 0.614 0.580 1.000 0.373 0.952 ORS844 9 94.9 0.074 1.000 HT645 10 21.1 0.221 0.551 0.241 0.492 0.051 0.207 HT347 10 23.9 0.237 0.673 HT692 10 24.4 0.034 0.459 ORS815 10 45.3 0.189 0.886 ORS1095 10 53.6 0.038 0.653 ORS613 10 54.3 0.094 0.760 ORS853 10 58.9 0.117 1.000 HT615 10 76.9 0.007 0.043 0.240 1.000 0.244 1.000 HT424 11 26.2 0.000 0.000 0.000 0.000 0.000 0.000 HT701 11 41.7 0.061 0.775 0.165 1.000 0.100 0.540 ORS1091 11 42.5 0.004 0.007 HT555 11 47.8 0.032 0.130 0.344 1.000 0.365 1.000 ORS697 11 49.0 0.041 0.796 HT732 11 55.4 0.102 0.689 0.342 1.000 0.117 0.364 HT815 12 0.0 0.028 0.261 HT882 12 18.5 0.015 0.117 0.121 0.464 0.186 0.723 ORS632 12 41.3 0.081 0.680 HT466 12 47.0 0.090 0.504 0.054 0.270 0.215 0.933 HT589 12 47.0 0.015 0.023 HT420 12 57.4 0.016 0.165 0.143 1.000 0.179 1.000 ORS200 13 0.6 0.167 0.761 HT772 13 0.6 0.019 0.170 HT854 13 26.0 0.119 0.864 0.053 0.237 0.171 0.760 HT739 13 30.1 0.229 1.000 0.547 1.000 0.542 1.000 HT356 13 30.1 0.099 0.275 0.020 0.048 0.152 0.437 HT848 13 30.1 0.039 0.204 0.487 1.000 0.552 1.000 HT295 13 31.4 0.051 0.179 HT540 13 38.9 0.055 0.193 0.124 0.476 0.220 0.875 ORS503 13 41.4 0.087 0.729 HT333 13 47.8 0.016 0.173 0.034 0.239 0.065 0.515 ORS799 13 52.9 0.012 0.116 HT382 13 68.9 0.114 0.899 0.245 1.000 0.241 1.000 ORS1086 14 31.7 0.027 0.333 HT621 14 31.7 0.017 0.075 0.288 1.000 0.246 0.907 HT429 14 31.7 0.299 1.000 0.304 1.000 0.012 0.032 HT765 14 42.6 0.072 0.518 0.345 0.725 0.279 0.680 HT979 14 52.6 0.093 0.749 0.072 0.431 0.184 1.000 HT329 15 23.8 0.039 0.726 0.092 1.000 0.112 1.000 HT985 15 41.5 0.217 0.674 0.732 1.000 0.460 1.000 HT716 15 57.3 0.153 0.750 0.480 1.000 0.472 1.000 HT451 15 86.0 0.042 0.095 0.155 0.232 0.083 0.122 HT813 15 89.7 0.037 0.648 0.132 1.000 0.150 1.000 HT953 15 90.9 0.057 0.171 0.439 0.813 0.257 0.477 HT283 15 92.1 0.238 0.774 0.291 0.909 0.012 0.031 HT279 16 5.3 0.008 0.060 0.070 0.362 0.052 0.237 HT323 16 11.0 0.093 0.270 0.579 1.000 0.313 0.981 HT439 16 12.6 0.029 0.141 0.155 0.781 0.275 1.000 ORS128 16 34.6 0.117 1.000 ORS1017 16 37.1 0.005 0.042 HT751 17 0.0 0.023 0.313 0.160 1.000 0.178 1.000 ORS565 17 13.8 0.020 0.401 ORS297 17 21.8 0.000 0.000 HT715 17 21.8 0.018 0.456 0.101 1.000 0.126 1.000 ORS169 17 21.8 0.069 0.839 ORS170 17 21.8 0.014 0.066 ORS634 17 33.8 0.013 0.185 ORS719 17 41.8 0.121 0.839 ORS847 17 41.8 0.058 0.252 ORS798 17 41.8 0.025 0.119 HT758 17 41.8 0.409 1.000 0.090 0.182 0.600 1.000 HT536 17 42.5 0.101 0.697 0.302 0.621 0.489 0.935 HT538 17 52.2 0.210 0.760 0.046 0.115 0.139 0.642 HT998 17 53.3 0.191 0.695 0.404 1.000 0.352 1.000 HT289 17 58.8 0.126 0.807 0.085 0.480 0.241 1.000 HT719 17 61.3 0.027 0.359 0.146 1.000 0.138 1.000 HT299 0.065 0.393 0.343 0.893 0.193 0.573 ORS125 0.019 0.104 ORS229 0.006 0.075 ORS285 0.024 0.521 ORS934 0.007 0.048 ORS976 0.016 0.665 ORS1088 0.011 0.267 ORS1152 0.053 0.710 ORS and HT indicate anonymous and EST-associated microsatellites, respectively. Markers whose names are underlined were used for the comparisons with H. argophyllus. Map Positions correspond to the map of the RHA801 x RHA280 population described in TANG et al. (2002). Positions in bold type were estimated from the NMS373 x ANN1811 population described in GANDHI et al. (2005), the HA370 x HA372 population described in GEDIL et al. (2001) or the PHA x PHB population described in YU et al. (2003). FST = was calculated according to WEIR and COCKERHAM
(1984), whereas G'ST is a standardized measure of differentiation developed by HEDRICK (2005). TABLE S2
Map positions, Gamma'ST, FST, and number of fixed differences for sequenced genes
Locus Linkage Linkage Map Fragment Gamma'ST FST Fixed Dif. Name Group Type Position size (bp) ann: pet ann: pet ann: pet HT206 1 C 12 203 0.347 0.465 0 HT083 3 C 23.1 190 0.385 0.508 1 HT205 10 C 10.7 189 0.222 0.298 0 HT250 10 C 26 188 0.475 0.605 2 HT179 10 C 49.9 153 0.23 0.314 0 HT229 10 C 32 305 0.376 0.501 1 HT047 13 R 1.2 317 0.404 0.525 1 HT196 14 R 9.3 418 0.162 0.208 0 HT065 14 R 61.6 240 0.056 0.023 0 HT163 16 R 19.4 277 0.198 0.254 0 HT037 16 R 32.7 396 0.505 0.635 4 HT052 16 R 69.9 250 0.286 0.382 1 HT064 17 R 13.8 266 0.125 0.145 0 HT254 17 R 39.2 493 0.486 0.615 4 Loci were mapped by LAI et al (2005) using the RHA801 x RHA280 population described in TANG et al. (2002). C = Collinear; R = Rearranged. Gamma'ST is a standardized measure of differentiation for nucleotide data ( HEDRICK 2005). FST = was calculated according to HUDSON et al. (1992).
LITERATURE CITED GANDHI, S. D., A. F. HEESACKER, C. A. FREEMAN, J. ARGYRIS, K. BRADFORD and S. J. KNAPP, 2005 The self-incompatibility locus (S) and quantitative trait loci for self- pollination and seed dormancy in sunflower. Theor. Appl. Genet. 111: 619-629. GEBIL, M. A., C. WYE, S. T. BERRY, B. SEGERS, J. PELEMAN et al., 2001 An integrated RFLP-AFLP linkage map for cultivated sunflower. Genome 44: 213-221. HEDRICK, P. W., 2005 A standardized genetic differentiation measure. Evolution 59: 1633-1638. HUDSON, R. R., M. SLATKIN, and W. P. MADDISON, 1992 Estimation of levels of gene flow from DNA-sequence data. Genetics 132: 583-589. LAI, Z., K. LIVINGSTONE, Y. ZOU, S. A. CHURCH, S. J. KNAPP, J. ANDREWS, and L. H. RIESEBERG. 2005 Identification and mapping of SNPs from ESTs in sunflower. Theor. Appl. Genet. 111: 1532-1544. TANG, S., J. K. YU, M. B. SLABAUGH, D. K. SHINTANI and S. J. KNAPP, 2002 Simple sequence repeat map of the sunflower genome. Theor. Appl. Genet. 105: 1124- 1136. WEIR, B. S., and C. C. COCKERHAM, 1984 Estimating F-statics for the analysis of population structure. Evolution 38: 1358-1370. YU, J. K., S. TANG, M. B. SLABAUGH, A. HEESACKER, G. COLE et al., 2003 Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci. 43: 367-387.