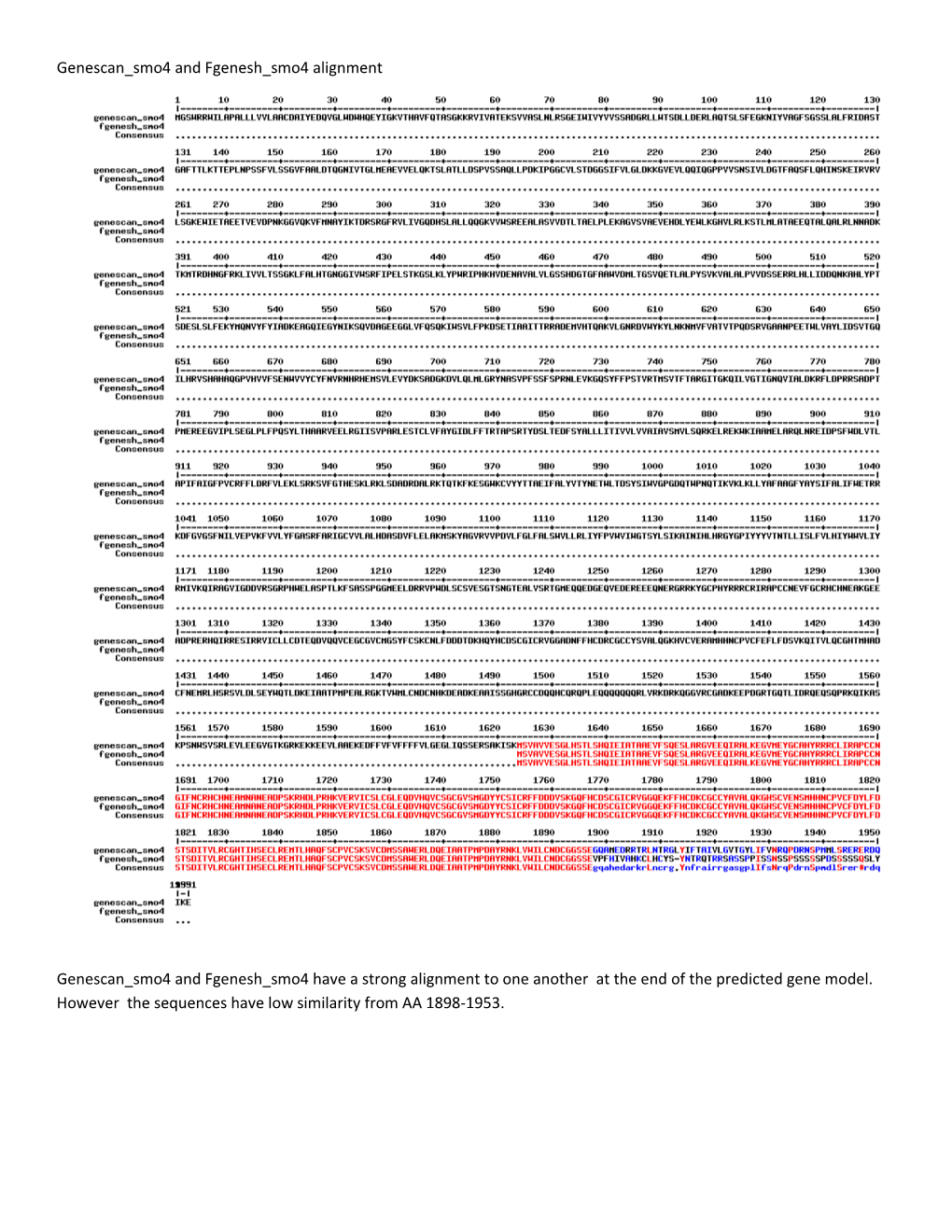
Genescan_smo4 and Fgenesh_smo4 alignment
Genescan_smo4 and Fgenesh_smo4 have a strong alignment to one another at the end of the predicted gene model. However the sequences have low similarity from AA 1898-1953. Multalign genescan_smo4 and Fgenesh_smo5
As with the alignment before, this alignment is fairly strong also. There are 6 main differences between the two sequences. It looks as though both gene models have additional exons (or extended certain exons) that the other gene model does not have. There are three regions where the sequence similarity is low, perhaps those portions of the sequence are in a different reading frame.
Blastp results for Genescan_smo4
Putative conserved domains have been detected, click on the image below for detailed results.
Best Blast hits
>ref|XP_001765108.1| predicted protein [Physcomitrella patens subsp. patens]
gb|EDQ70103.1| predicted protein [Physcomitrella patens subsp. patens] Length=998
GENE ID: 5928285 PHYPADRAFT_184382 | hypothetical protein [Physcomitrella patens subsp. patens] (10 or fewer PubMed links)
Score = 850 bits (2196), Expect = 0.0, Method: Compositional matrix adjust. Identities = 468/990 (47%), Positives = 651/990 (65%), Gaps = 121/990 (12%)
Query 14 LLVVLA----ACDAIYEDQVGLWDWHQEYIGKVTHAVFQT-ASGKKRVIVATEKSVVASL 68 L VV+A C A+YEDQVG+ DWHQ+YIG+V HAVFQT +G+KRV+VATE++ +ASL Sbjct 13 LFVVVACLSSTCLALYEDQVGVRDWHQQYIGRVKHAVFQTQGTGRKRVVVATEQNAIASL 72
Query 69 NLRSGEIWIVYVVSSA------DGRLLWTSDL----- 94 NLR+G+I+ +V+ DG L+W + + Sbjct 73 NLRTGDIYWRHVLGETDNIDALEISMGKYVLTLSKGNTVRAWHLPDGALIWETRIQAFQG 132
Query 95 LDERLAQTSLSFEG---KNIYVAGFSGSSL------ALFRIDA--STGAFTTLKTTE 140 + L + + +G +++V +SGS L L+R+DA S F Sbjct 133 FNLGLIKLPVDIDGDKVNDLFV--YSGSILTAISGADGATLWRVDAAGSKNIFIEKVVLA 190
Query 141 PLNPSSFVLS----SGV-FAALDTQGNIVTGLMEAEV------VELQKTS 179 P ++ L GV +D + ++ L AE+ V LQ + Sbjct 191 PEEGKAYGLGFFGIMGVALVEIDLKTGDLSDLKSAELSSMLSTEHLHVTSDYAVALQSDA 250
Query 180 LATLLDSPVSSAQLLPDKIPGGCVLSTDGGSIFVLGLDKKGV------221 + ++ S +L+ + P +L+ G S+ +L + +GV Sbjct 251 ESLVVALINSHKELIVVETPVSSILTNPGTSLKLLSTNLEGVISLSSDDQTVILKVDPTT 310
Query 222 ---EVLQQIQGPPVVSNSI-VLDGTFAQSFLQHINSKE------IRVRVLSGKEWIETAE 271 +++++ G VS+S+ VLD +A + ++ S+E +RV E + Sbjct 311 GKLSLVERLTGAVAVSDSLSVLDDKYATAIVEF--SEEGSAQNVFNLRVKGNDFSDEVQK 368
Query 272 ETVEVDPNKGGVQKVFMNAYIKTDRSRGFRVLIVGQDHSLALLQQGKVVWSREEALASVV 331 ETV++ ++G +QK F+NAY++TDRS GFR L+VG+D SL+LLQQG+VVW+RE+ LAS+V Sbjct 369 ETVKLPSHRGFIQKAFLNAYVRTDRSHGFRALVVGEDDSLSLLQQGEVVWTREDGLASIV 428
Query 332 DTLTAELPLEKAGVSVAEVEHDLYEWLKGHVLRLKSTLMLATAEEQTALQALRLNNADKT 391 D AELPLEK GVSVAEVEHDL EWLKGH++++K+TL LAT +E A+Q RLN ADKT Sbjct 429 DASPAELPLEKDGVSVAEVEHDLAEWLKGHIMKMKATLFLATPDELAAVQRARLNQADKT 488
Query 392 KMTRDHNGFRKLIVVLTSSGKLFALHTGNGGIVWSRFIPELSTK------GSLKLYPWRI 445 K TRDHNGFRKL+VVLT +GK+ ALHTG+G +VWS +P L LK++ W++ Sbjct 489 KHTRDHNGFRKLLVVLTKAGKISALHTGDGHVVWSLLVPSLRASYGNPRFSPLKIFQWQV 548
Query 446 PHKH-VDENAVALVLGSS---HDGTGFAAWVDMLTGSVQETLALPYSVKVALALPVVDSS 501 PH+H +DEN V L+L + +D G +W+D+ G+ +++ L YSV + PV DSS Sbjct 549 PHQHALDENPVVLILAQADPGYDVKGALSWIDVHKGTELQSVKLSYSVTQVVTTPVTDSS 608
Query 502 ERRLHLLIDDQNKAHLYPTSDESLSLFEKYMQNVYFYIADKEAGQIEGYNIKSQVDAGE- 560 E+RLHLLID++ +AHL+P ++ESL+LF KY +N YFY DK ++ GY + VD Sbjct 609 EQRLHLLIDNRKRAHLFPATEESLALFLKYKENAYFYEVDKADQKMHGYGLLDLVDPSTG 668
Query 561 --EGGLVFQSQKIWSVLFPKDSETIAAITTRRADEMVHTQAKVLGNRDVWYKYLNKNMVF 618 + G VF+S+K+WS++FP ++E+I + TR++DE+ HTQ KVL NRD+ +KYLNKN+VF Sbjct 669 NIKEGYVFESRKLWSIVFPAETESITTVVTRKSDEVTHTQTKVLSNRDILFKYLNKNLVF 728
Query 619 VATVTPQD-SRVGAANPEETWLVAYLIDSVTGQILHRVSHAHAQGPVHVVFSENWVVYCY 677 VATV P+D S+VGA +PEE LV YL+D+VTG+ILHRVSH + QGPVH V SENWVVY Y Sbjct 729 VATVAPKDKSQVGAVSPEEKTLVVYLVDTVTGRILHRVSHPNMQGPVHAVLSENWVVYHY 788
Query 678 FNVRNHRHEMSVLEVYDKS-ADGKDVLQLMLGRYNASVPFSSFSPRNLEVKGQSYFFPST 736 FN+R HR+EMSVLE+YD+S K V+QLMLG++N+SVP SS+SP NLEVK QSYFF T Sbjct 789 FNLRQHRYEMSVLEIYDQSRLPDKGVIQLMLGQHNSSVPISSYSPVNLEVKQQSYFFTFT 848
Query 737 VRTMSVTFTARGITGKQILVGTIGNQVIALDKRFLDPRRSADPTPMEREEGVIPLSEGLP 796 V+TM+VT TA+GIT KQ+L+GT+ +QV+ALDKR DPRR+ PTP E+EEG++PL++ +P Sbjct 849 VKTMTVTSTAKGITAKQLLLGTVNDQVLALDKRLFDPRRTLTPTPAEQEEGILPLTDSIP 908
Query 797 LFPQSYLTHAARVEELRGIISVPARLESTCLVFAYGIDLFFTRTAPSRTYDSLTEDFSYA 856 + PQSYLTH+ +VE LRG++++PARLEST LVFAYG+DLF+T TAPS+ YDSLTEDFSYA Sbjct 909 ISPQSYLTHSYQVEGLRGLLTIPARLESTSLVFAYGLDLFYTHTAPSKIYDSLTEDFSYA 968
Query 857 LLLITIVVLVVAIAVSMVLSQRKELREKWK 886 LLL+TIVVL ++I V+ VLS+R+EL EKWK Sbjct 969 LLLVTIVVLFLSIIVTYVLSERRELAEKWK 998
>emb|CAO44049.1| unnamed protein product [Vitis vinifera] Length=987
Score = 800 bits (2065), Expect = 0.0, Method: Compositional matrix adjust. Identities = 433/968 (44%), Positives = 615/968 (63%), Gaps = 108/968 (11%)
Query 23 AIYEDQVGLWDWHQEYIGKVTHAVFQT------49 ++YEDQVGL DWHQ+YIGKV HAVF T Sbjct 24 SLYEDQVGLMDWHQQYIGKVKHAVFHTQKAGRKRVVVSTEENVIASLDLRRGDIFWRHVL 83
Query 50 ------ASGKKRVIVATEKSVVASLNLRSGE-IW------76 A GK + +++E S++ + NL G+ +W Sbjct 84 GPNDAVDEIDIALGKYVITLSSEGSILRAWNLPDGQMVWESFLQGPKPSKSLLSVSANLK 143
Query 77 -----IVYV------VSSADGRLLWTSDLLDERL--AQTSLSFEGKNIYVAGFSG-SS 120 +++V VSS DG +LW D DE L Q IY GF G S Sbjct 144 IDKDNVIFVFGKGCLHAVSSIDGEVLWKKDFADESLEVQQIIHPLGSDMIYAVGFVGLSQ 203
Query 121 LALFRIDASTGAFTTLKTTEPLNPSSF-----VLSSGVFAALD-TQGNIVT-GLMEAEVV 173 L ++I+ G LK P F ++SS ALD T+ ++++ ++ E+ Sbjct 204 LDAYQINVRNGE--VLKHRSAAFPGGFCGEVSLVSSDTLVALDATRSSLISISFLDGEI- 260
Query 174 ELQKTSLATLLDSPVSSAQLLPDKIPGGCVLSTDGGSIFVLGLDKKGVEVLQQIQGPPVV 233 LQ+T ++ L+ A +LP K+ G ++ D +FV D+ +EV ++I Sbjct 261 SLQQTHISNLVGDSFGMAVMLPSKLSGMLMIKIDNYMVFVRVADEGKLEVAEKINDAAAA 320
Query 234 SNSIVLDGTFAQSF--LQHINSKEIRVRVLSGKEWI-ETAEETVEVDPNKGGVQKVFMNA 290 + + Q+F ++H +K I + V +W + +E++ +D +G V K+F+N+ Sbjct 321 VSDALALSEGQQAFGLVEHGGNK-IHLTVKLVNDWNGDLLKESIRMDHQRGCVHKIFINS 379
Query 291 YIKTDRSRGFRVLIVGQDHSLALLQQGKVVWSREEALASVVDTLTAELPLEKAGVSVAEV 350 YI+TDRS GFR LIV +DHSL LLQQG++VWSRE+ LAS++D +ELP+EK GVSVA+V Sbjct 380 YIRTDRSHGFRALIVMEDHSLLLLQQGEIVWSREDGLASIIDVTASELPVEKEGVSVAKV 439
Query 351 EHDLYEWLKGHVLRLKSTLMLATAEEQTALQALRLNNADKTKMTRDHNGFRKLIVVLTSS 410 EH+L+EWLKGH+L+LK TLMLA+ E+ A+Q +RL +++K+KMTRDHNGFRKL++VLT + Sbjct 440 EHNLFEWLKGHMLKLKGTLMLASPEDMIAIQGMRLKSSEKSKMTRDHNGFRKLLIVLTRA 499
Query 411 GKLFALHTGNGGIVWSRFIPELSTKGS------LKLYPWRIPHKH-VDENAVALVLGS-- 461 GKLFALHTG+G +VWS + L + L +Y W++PH H +DEN LV+G Sbjct 500 GKLFALHTGDGRVVWSVLLHSLHNSEACAYPTGLNVYQWQVPHHHAMDENPSVLVVGRCG 559
Query 462 -SHDGTGFAAWVDMLTGSVQETLALPYSVKVALALPVVDSSERRLHLLIDDQNKAHLYPT 520 D G ++VD TG ++L L +S++ + L DS E+RLHL+ID + AHLYP Sbjct 560 LGSDAPGVLSFVDTYTGKELDSLFLTHSIERIIPLSFTDSREQRLHLIIDTDHHAHLYPR 619
Query 521 SDESLSLFEKYMQNVYFYIADKEAGQIEGYNIKSQVDAGEEGGLVFQSQKIWSVLFPKDS 580 + E++ +F+ + N+Y+Y + E G I G+ +KS E F ++ +WS++FP +S Sbjct 620 TPEAIGIFQHELPNIYWYSVEAENGIIRGHALKSNCILQEGDEYCFDTRDLWSIVFPSES 679
Query 581 ETIAAITTRRADEMVHTQAKVLGNRDVWYKYLNKNMVFVATVTPQDS-RVGAANPEETWL 639 E I A TR+ +E+VHTQAKV+ ++DV YKY++KN++FVATV P+ + +G+ PEE+WL Sbjct 680 EKILATVTRKLNEVVHTQAKVITDQDVMYKYVSKNLLFVATVAPKATGEIGSVTPEESWL 739
Query 640 VAYLIDSVTGQILHRVSHAHAQGPVHVVFSENWVVYCYFNVRNHRHEMSVLEVYDKS-AD 698 V YLID+VTG+I++R++H QGPVH VFSENWVVY YFN+R HR+EMSV+E+YD+S AD Sbjct 740 VVYLIDTVTGRIIYRMTHHGTQGPVHAVFSENWVVYHYFNLRAHRYEMSVVEIYDQSRAD 799
Query 699 GKDVLQLMLGRYNASVPFSSFSPRNLEVKGQSYFFPSTVRTMSVTFTARGITGKQILVGT 758 KDV +L+LG++N + P SS+S + K Q YFF +V+ M+VT TA+GIT KQ+L+GT Sbjct 800 NKDVWKLVLGKHNLTSPVSSYSRPEVITKSQFYFFTHSVKAMAVTSTAKGITSKQLLIGT 859
Query 759 IGNQVIALDKRFLDPRRSADPTPMEREEGVIPLSEGLPLFPQSYLTHAARVEELRGIISV 818 IG+QV+ALDKR+LDPRR+ +P+ EREEG+IPL++ LP+ PQSY+TH +VE LRGI++ Sbjct 860 IGDQVLALDKRYLDPRRTINPSQSEREEGIIPLTDSLPIIPQSYVTHNLKVEGLRGIVTA 919
Query 819 PARLESTCLVFAYGIDLFFTRTAPSRTYDSLTEDFSYALLLITIVVLVVAIAVSMVLSQR 878 PA+LEST LVFAYG+DLFFTR APSRTYD LT+DFSYALLLITIV LV AI V+ +LS+R Sbjct 920 PAKLESTTLVFAYGVDLFFTRIAPSRTYDLLTDDFSYALLLITIVALVAAIFVTWILSER 979 Query 879 KELREKWK 886 KEL+EKW+ Sbjct 980 KELQEKWR 987
>ref|XP_001772470.1| predicted protein [Physcomitrella patens subsp. patens]
gb|EDQ62752.1| predicted protein [Physcomitrella patens subsp. patens] Length=252 /note="CHY zinc finger. This family of domains are likely
to bind to zinc ions. They contain many conserved cysteine and histidine residues. We have named this domain after the N-terminal motif CXHY. This domain can be found in isolation in some proteins, but...; cl01802"
GENE ID: 5935675 PHYPADRAFT_138830 | hypothetical protein [Physcomitrella patens subsp. patens] (10 or fewer PubMed links)
Score = 322 bits (824), Expect = 3e-85, Method: Compositional matrix adjust. Identities = 141/223 (63%), Positives = 180/223 (80%), Gaps = 1/223 (0%)
Query 1675 CAHYRRRCLIRAPCCNGIFNCRHCHNEAMNANEADPSKRHDLPRHKVERVICSLCGLEQD 1734 CAHY+R C IRAPCCN +F+CRHCHN+A + NE D ++RH++ R VE+VICSLC EQD Sbjct 1 CAHYKRGCKIRAPCCNEVFDCRHCHNDAKSVNEKDDTQRHEIDRRLVEKVICSLCDHEQD 60
Query 1735 VHQVCSGCGVSMGDYYCSICRFFDDDVSKGQFHCDSCGICRVGGQEKFFHCDKCGCCYAV 1794 V QVC CGV MG+Y+CS C+FFDDD SK QFHCD CGICR+GG++ FFHCD+CGCCY+V Sbjct 61 VQQVCENCGVCMGEYFCSKCKFFDDDTSKRQFHCDKCGICRIGGRDNFFHCDRCGCCYSV 120
Query 1795 ALQKGHSCVENSMHHNCPVCFDYLFDSTSDITVLRCGHTIHSECLREMTLHAQFSCPVCS 1854 L++ H+CVE SMH +C +C +YLFDS DITVL CGHT+H ECL+EM H Q++CP+C+ Sbjct 121 ELRERHTCVEKSMHQDCAICMEYLFDSLMDITVLPCGHTLHLECLQEMYKHYQYNCPLCN 180
Query 1855 KSVCDMSSAWERLDQEIAATPMPDAYRNKLVWILCNDCGGSSE 1897 KSVCDMSS W+ +D EIA+ MP+ ++++VWILCNDCG +E Sbjct 181 KSVCDMSSVWKEIDLEIASIQMPEN-QSRMVWILCNDCGAKNE 222
Score = 295 bits (756), Expect = 2e-77, Method: Compositional matrix adjust. Identities = 136/223 (60%), Positives = 165/223 (73%), Gaps = 10/223 (4%)
Query 1269 CPHYRRRCRIRAPCCNEVFGCRHCHNEAKG-EEADPRERHQIRRESIRRVICLLCDTEQD 1327 C HY+R C+IRAPCCNEVF CRHCHN+AK E D +RH+I R + +VIC LCD EQD Sbjct 1 CAHYKRGCKIRAPCCNEVFDCRHCHNDAKSVNEKDDTQRHEIDRRLVEKVICSLCDHEQD 60
Query 1328 VQQVCEGCGVCMGSYFCSKCNLFDDDTDKHQYHCDSCGICRVGGADNFFHCDRCGCCYSV 1387 VQQVCE CGVCMG YFCSKC FDDDT K Q+HCD CGICR+GG DNFFHCDRCGCCYSV Sbjct 61 VQQVCENCGVCMGEYFCSKCKFFDDDTSKRQFHCDKCGICRIGGRDNFFHCDRCGCCYSV 120
Query 1388 ALQGKHVCVERAMHHNCPVCFEFLFDSVKQITVLQCGHTMHADCFNEMRLH------S 1439 L+ +H CVE++MH +C +C E+LFDS+ ITVL CGHT+H +C EM H + Sbjct 121 ELRERHTCVEKSMHQDCAICMEYLFDSLMDITVLPCGHTLHLECLQEMYKHYQYNCPLCN 180
Query 1440 RSVLDLSEYWQTLDKEIAATPMPEALRGKTVWMLCNDCNHKDE 1482 +SV D+S W+ +D EIA+ MPE + + VW+LCNDC K+E Sbjct 181 KSVCDMSSVWKEIDLEIASIQMPEN-QSRMVWILCNDCGAKNE 222 TAIR database results
Score E Sequences producing significant alignments: (bits) Value ref|NP_196717.3| catalytic [Arabidopsis thaliana] 747 0.0 ref|NP_197938.2| zinc finger (C3HC4-type RING finger) famil... 375 e-103 ref|NP_197683.1| zinc finger (C3HC4-type RING finger) famil... 367 e-101 ref|NP_001078621.1| zinc finger (C3HC4-type RING finger) fa... 358 2e-98 ref|NP_197366.1| zinc finger (C3HC4-type RING finger) famil... 338 3e-92 ref|NP_191856.4| protein binding / zinc ion binding [Arabid... 316 1e-85 ref|NP_001031037.1| LAG13 (LAG1 LONGEVITY ASSURANCE HOMOLOG... 301 2e-81 ref|NP_566769.1| LAG1 (Longevity assurance gene 1) [Arabido... 290 7e-78 ref|NP_172815.2| LAG13 (LAG1 LONGEVITY ASSURANCE HOMOLOG 3)... 236 2e-61 ref|NP_177615.2| protein binding / zinc ion binding [Arabid... 202 2e-51 ref|NP_188457.1| EMB2454 (EMBRYO DEFECTIVE 2454); protein b... 191 4e-48 ref|NP_188557.1| LAG1 HOMOLOG 2 (LONGEVITY ASSURANCE GENE1 ... 191 5e-48 ref|NP_173325.2| protein binding / zinc ion binding [Arabid... 190 7e-48 ref|NP_566651.1| zinc finger (C3HC4-type RING finger) famil... 40 0.018 ref|NP_565253.1| RHA2B (RING-H2 FINGER PROTEIN 2B); protein... 39 0.024 ref|NP_191705.1| BRH1 (BRASSINOSTEROID-RESPONSIVE RING-H2);... 39 0.025 ref|NP_178507.1| XERICO; protein binding / zinc ion binding... 39 0.026 ref|NP_973416.1| XERICO; protein binding / zinc ion binding... 39 0.026 ref|NP_567480.2| zinc finger (C3HC4-type RING finger) famil... 39 0.032 ref|NP_188629.1| zinc finger (C3HC4-type RING finger) famil... 39 0.037 ref|NP_177367.1| zinc finger (C3HC4-type RING finger) famil... 39 0.047 ref|NP_974274.1| zinc finger (C3HC4-type RING finger) famil... 39 0.047 ref|NP_188049.1| zinc finger (C3HC4-type RING finger) famil... 38 0.055 ref|NP_565942.1| RHC1A (RING-H2 finger C1A); protein bindin... 38 0.066 ref|NP_973651.1| RHC1A (RING-H2 finger C1A); protein bindin... 38 0.066 ref|NP_973652.1| RHC1A (RING-H2 finger C1A); protein bindin... 38 0.066
>ref|NP_196717.3| catalytic [Arabidopsis thaliana] Length = 982 /note="Dehydrogenases with pyrrolo-quinoline quinone (PQQ)
as cofactor, like ethanol, methanol, and membrane bound glucose dehydrogenases. The alignment model contains an 8-bladed beta-propeller; cl09980"
Score = 747 bits (1928), Expect = 0.0, Method: Composition-based stats. Identities = 409/967 (42%), Positives = 594/967 (61%), Gaps = 108/967 (11%)
Query: 23 AIYEDQVGLWDWHQEYIGKVTHAVFQT------49 ++YEDQ GL DWHQ YIGKV HAVF T Sbjct: 21 SLYEDQAGLTDWHQRYIGKVKHAVFHTQKTGRKRVIVSTEENVVASLDLRHGEIFWRHVL 80
Query: 50 ------ASGKKRVIVATEKSVVASLNLRSGE-IW------76 A GK + +++E S + + NL G+ +W Sbjct: 81 GTKDAIDGVGIALGKYVITLSSEGSTLRAWNLPDGQMVWETSLHTAQHSKSLLSVPINLK 140
Query: 77 ------IVYVVSSADGRLLWTSDLLDERL-AQTSLSFEGKNI-YVAGFSGSSL 121 ++ VS+ DG +LW D E Q L G +I YV GF SS Sbjct: 141 VDKDYPITVFGGGYLHAVSAIDGEVLWKKDFTAEGFEVQRVLQAPGSSIIYVLGFLHSSE 200
Query: 122 AL-FRIDASTGAFTTLKTTEPLNPSSF-----VLSSGVFAALDTQGNIVT--GLMEAEVV 173 A+ ++ID+ +G K+T + P F +SS LD+ +I+ G ++ ++ Sbjct: 201 AVVYQIDSKSGEVVAQKST--VFPGGFSGEISSVSSDKVVVLDSTRSILVTIGFIDGDI- 257 Query: 174 ELQKTSLATLLDSPVSSAQLLPDKIPGGCVLSTDGGSIFVLGLDKKGVEVLQQIQGPPVV 233 QKT ++ L++ +A++L + + + +IFV DK +EV+ + + Sbjct: 258 SFQKTPISDLVEDS-GTAEILSPLLSNMLAVKVNKRTIFVNVGDKGKLEVVDSLSDETAM 316
Query: 234 SNSI-VLDGTFAQSFLQHINSK-EIRVRVLSGKEWIETAEETVEVDPNKGGVQKVFMNAY 291 S+S+ V D A + + H S+ + V++++ + ET+++D N+G V KVFMN Y Sbjct: 317 SDSLPVADDQEAFASVHHEGSRIHLMVKLVNDLNNV-LLRETIQMDQNRGRVHKVFMNNY 375
Query: 292 IKTDRSRGFRVLIVGQDHSLALLQQGKVVWSREEALASVVDTLTAELPLEKAGVSVAEVE 351 I+TDRS GFR LIV +DHSL LLQQG +VWSREE LASV D TAELPLEK GVSVA+VE Sbjct: 376 IRTDRSNGFRALIVMEDHSLLLLQQGAIVWSREEGLASVTDVTTAELPLEKDGVSVAKVE 435
Query: 352 HDLYEWLKGHVLRLKSTLMLATAEEQTALQALRLNNADKTKMTRDHNGFRKLIVVLTSSG 411 H L+EWLKGHVL+LK +L+LA+ E+ A+Q LR+ ++ K K+TRDHNGFRKLI+ LT +G Sbjct: 436 HTLFEWLKGHVLKLKGSLLLASPEDVVAIQDLRVKSSGKNKLTRDHNGFRKLILALTRAG 495
Query: 412 KLFALHTGNGGIVWSRFIPELSTKGS------LKLYPWRIPHKH-VDENAVALVL---GS 461 KLFALHTG+G IVWS + S S + LY W++PH H +DEN LV+ GS Sbjct: 496 KLFALHTGDGRIVWSMLLNSPSQSQSCERPNGVSLYQWQVPHHHAMDENPSVLVVGKCGS 555
Query: 462 SHDGTGFAAWVDMLTGSVQETLALPYSVKVALALPVVDSSERRLHLLIDDQNKAHLYPTS 521 G ++VD+ TG + + +SV + LP+ DS E+RLHL+ D HLYP + Sbjct: 556 DSSAPGVLSFVDVYTGKEISSSDIGHSVVQVMPLPITDSKEQRLHLIADTVGHVHLYPKT 615
Query: 522 DESLSLFEKYMQNVYFYIADKEAGQIEGYNIKSQVDAGEEGGLVFQSQKIWSVLFPKDSE 581 E+LS+F++ QNVY+Y + + G I G+ +K F ++++W+V+FP +SE Sbjct: 616 SEALSIFQREFQNVYWYTVEADDGIIRGHVMKGSCSGETADEYCFTTRELWTVVFPSESE 675
Query: 582 TIAAITTRRADEMVHTQAKVLGNRDVWYKYLNKNMVFVATVTPQDS-RVGAANPEETWLV 640 I + TR+ +E+VHTQAKV ++D+ YKY+++N++FVATV+P+ + +G+ PEE+ LV Sbjct: 676 KIISTLTRKPNEVVHTQAKVNTDQDLLYKYVSRNLLFVATVSPKGAGEIGSVTPEESSLV 735
Query: 641 AYLIDSVTGQILHRVSHAHAQGPVHVVFSENWVVYCYFNVRNHRHEMSVLEVYDKS-ADG 699 YLID++TG+ILHR+SH QGPVH VFSENWVVY YFN+R H++E++V+E+YD+S A+ Sbjct: 736 VYLIDTITGRILHRLSHQGCQGPVHAVFSENWVVYHYFNLRAHKYEVTVVEIYDQSRAEN 795
Query: 700 KDVLQLMLGRYNASVPFSSFSPRNLEVKGQSYFFPSTVRTMSVTFTARGITGKQILVGTI 759 K+V +L+LG++N + P +S+S + K QSYFF +V+T++VT TA+GIT KQ+L+GTI Sbjct: 796 KNVWKLILGKHNLTAPITSYSRPEVFTKSQSYFFAQSVKTIAVTSTAKGITSKQLLIGTI 855
Query: 760 GNQVIALDKRFLDPRRSADPTPMEREEGVIPLSEGLPLFPQSYLTHAARVEELRGIISVP 819 G+Q++ALDKRF+DPRR+ +P+ E+EEG+IPL++ LP+ PQ+Y+TH+ +VE LRGI++ P Sbjct: 856 GDQILALDKRFVDPRRTLNPSQAEKEEGIIPLTDTLPIIPQAYVTHSHKVEGLRGIVTAP 915
Query: 820 ARLESTCLVFAYGIDLFFTRTAPSRTYDSLTEDFSYXXXXXXXXXXXXXXXXXXXXSQRK 879 ++LEST VFAYG+DLF+TR APS+TYDSLT+DFSY S++K Sbjct: 916 SKLESTTHVFAYGVDLFYTRLAPSKTYDSLTDDFSYALLLITIVALVAAIYITWVLSEKK 975
Query: 880 ELREKWK 886 EL EKW+ Sbjct: 976 ELSEKWR 982
>ref|NP_197938.2| zinc finger (C3HC4-type RING finger) family protein [Arabidopsis thaliana] Length = 308
Score = 375 bits (962), Expect = e-103, Method: Composition-based stats. Identities = 160/272 (58%), Positives = 199/272 (73%), Gaps = 5/272 (1%) Query: 1630 ESGLHSTLSHQIEIATAAEVFSQESLARGVEEQI--RALKEGVMEYGCAHYRRRCLIRAP 1687 E G S SH I +E +L R E + + L G+MEYGC HYRRRC IRAP Sbjct: 19 EKGEMSRHSHPHSINEESE---SSTLERVAAESLTNKVLDRGLMEYGCPHYRRRCCIRAP 75
Query: 1688 CCNGIFNCRHCHNEAMNANEADPSKRHDLPRHKVERVICSLCGLEQDVHQVCSGCGVSMG 1747 CCN IF C HCH EA N D +RHD+PRH+VE+VIC LCG EQ+V Q+C CGV MG Sbjct: 76 CCNEIFGCHHCHYEAKNNINVDQKQRHDIPRHQVEQVICLLCGTEQEVGQICIHCGVCMG 135
Query: 1748 DYYCSICRFFDDDVSKGQFHCDSCGICRVGGQEKFFHCDKCGCCYAVALQKGHSCVENSM 1807 Y+C +C+ +DDD SK Q+HCD CGICR+GG+E FFHC KCGCCY++ L+ GH CVE +M Sbjct: 136 KYFCKVCKLYDDDTSKKQYHCDGCGICRIGGRENFFHCYKCGCCYSILLKNGHPCVEGAM 195
Query: 1808 HHNCPVCFDYLFDSTSDITVLRCGHTIHSECLREMTLHAQFSCPVCSKSVCDMSSAWERL 1867 HH+CP+CF++LF+S +D+TVL CGHTIH +CL EM H Q++CP+CSKSVCDMS WE+ Sbjct: 196 HHDCPICFEFLFESRNDVTVLPCGHTIHQKCLEEMRDHYQYACPLCSKSVCDMSKVWEKF 255
Query: 1868 DQEIAATPMPDAYRNKLVWILCNDCGGSSEGQ 1899 D EIAATPMP+ Y+N++V ILCNDCG +E Q Sbjct: 256 DMEIAATPMPEPYQNRMVQILCNDCGKKAEVQ 287
Score = 323 bits (829), Expect = 5e-88, Method: Composition-based stats. Identities = 138/226 (61%), Positives = 164/226 (72%), Gaps = 9/226 (3%)
Query: 1266 KYGCPHYRRRCRIRAPCCNEVFGCRHCHNEAKGE-EADPRERHQIRRESIRRVICLLCDT 1324 +YGCPHYRRRC IRAPCCNE+FGC HCH EAK D ++RH I R + +VICLLC T Sbjct: 60 EYGCPHYRRRCCIRAPCCNEIFGCHHCHYEAKNNINVDQKQRHDIPRHQVEQVICLLCGT 119
Query: 1325 EQDVQQVCEGCGVCMGSYFCSKCNLFDDDTDKHQYHCDSCGICRVGGADNFFHCDRCGCC 1384 EQ+V Q+C CGVCMG YFC C L+DDDT K QYHCD CGICR+GG +NFFHC +CGCC Sbjct: 120 EQEVGQICIHCGVCMGKYFCKVCKLYDDDTSKKQYHCDGCGICRIGGRENFFHCYKCGCC 179
Query: 1385 YSVALQGKHVCVERAMHHNCPVCFEFLFDSVKQITVLQCGHTMHADCFNEMRLH------1438 YS+ L+ H CVE AMHH+CP+CFEFLF+S +TVL CGHT+H C EMR H Sbjct: 180 YSILLKNGHPCVEGAMHHDCPICFEFLFESRNDVTVLPCGHTIHQKCLEEMRDHYQYACP 239
Query: 1439 --SRSVLDLSEYWQTLDKEIAATPMPEALRGKTVWMLCNDCNHKDE 1482 S+SV D+S+ W+ D EIAATPMPE + + V +LCNDC K E Sbjct: 240 LCSKSVCDMSKVWEKFDMEIAATPMPEPYQNRMVQILCNDCGKKAE 285
>ref|NP_197683.1| zinc finger (C3HC4-type RING finger) family protein [Arabidopsis thaliana] Length = 291
Score = 367 bits (943), Expect = e-101, Method: Composition-based stats. Identities = 149/228 (65%), Positives = 184/228 (80%)
Query: 1669 GVMEYGCAHYRRRCLIRAPCCNGIFNCRHCHNEAMNANEADPSKRHDLPRHKVERVICSL 1728 G YGC+HYRRRC IRAPCC+ IF+CRHCHNEA ++ + RH+LPRH+V +VICSL Sbjct: 21 GSGHYGCSHYRRRCKIRAPCCDEIFDCRHCHNEAKDSLHIEQHHRHELPRHEVSKVICSL 80
Query: 1729 CGLEQDVHQVCSGCGVSMGDYYCSICRFFDDDVSKGQFHCDSCGICRVGGQEKFFHCDKC 1788 C EQDV Q CS CGV MG Y+CS C+FFDDD+SK Q+HCD CGICR GG+E FFHC +C Sbjct: 81 CETEQDVQQNCSNCGVCMGKYFCSKCKFFDDDLSKKQYHCDECGICRTGGEENFFHCKRC 140
Query: 1789 GCCYAVALQKGHSCVENSMHHNCPVCFDYLFDSTSDITVLRCGHTIHSECLREMTLHAQF 1848 CCY+ ++ H CVE +MHHNCPVCF+YLFDST DITVLRCGHT+H EC ++M LH ++ Sbjct: 141 RCCYSKIMEDKHQCVEGAMHHNCPVCFEYLFDSTRDITVLRCGHTMHLECTKDMGLHNRY 200
Query: 1849 SCPVCSKSVCDMSSAWERLDQEIAATPMPDAYRNKLVWILCNDCGGSS 1896 +CPVCSKS+CDMS+ W++LD+E+AA PMP Y NK+VWILCNDCG ++ Sbjct: 201 TCPVCSKSICDMSNLWKKLDEEVAAYPMPKMYENKMVWILCNDCGSNT 248
Score = 325 bits (832), Expect = 3e-88, Method: Composition-based stats. Identities = 137/220 (62%), Positives = 162/220 (73%), Gaps = 9/220 (4%)
Query: 1267 YGCPHYRRRCRIRAPCCNEVFGCRHCHNEAKGE-EADPRERHQIRRESIRRVICLLCDTE 1325 YGC HYRRRC+IRAPCC+E+F CRHCHNEAK + RH++ R + +VIC LC+TE Sbjct: 25 YGCSHYRRRCKIRAPCCDEIFDCRHCHNEAKDSLHIEQHHRHELPRHEVSKVICSLCETE 84
Query: 1326 QDVQQVCEGCGVCMGSYFCSKCNLFDDDTDKHQYHCDSCGICRVGGADNFFHCDRCGCCY 1385 QDVQQ C CGVCMG YFCSKC FDDD K QYHCD CGICR GG +NFFHC RC CCY Sbjct: 85 QDVQQNCSNCGVCMGKYFCSKCKFFDDDLSKKQYHCDECGICRTGGEENFFHCKRCRCCY 144
Query: 1386 SVALQGKHVCVERAMHHNCPVCFEFLFDSVKQITVLQCGHTMHADCFNEMRLHSR----- 1440 S ++ KH CVE AMHHNCPVCFE+LFDS + ITVL+CGHTMH +C +M LH+R Sbjct: 145 SKIMEDKHQCVEGAMHHNCPVCFEYLFDSTRDITVLRCGHTMHLECTKDMGLHNRYTCPV 204
Query: 1441 ---SVLDLSEYWQTLDKEIAATPMPEALRGKTVWMLCNDC 1477 S+ D+S W+ LD+E+AA PMP+ K VW+LCNDC Sbjct: 205 CSKSICDMSNLWKKLDEEVAAYPMPKMYENKMVWILCNDC 244
>ref|NP_001078621.1| zinc finger (C3HC4-type RING finger) family protein [Arabidopsis thaliana] Length = 328
Score = 358 bits (919), Expect = 2e-98, Method: Composition-based stats. Identities = 151/258 (58%), Positives = 189/258 (73%), Gaps = 5/258 (1%)
Query: 1630 ESGLHSTLSHQIEIATAAEVFSQESLARGVEEQI--RALKEGVMEYGCAHYRRRCLIRAP 1687 E G S SH I +E +L R E + + L G+MEYGC HYRRRC IRAP Sbjct: 19 EKGEMSRHSHPHSINEESE---SSTLERVAAESLTNKVLDRGLMEYGCPHYRRRCCIRAP 75
Query: 1688 CCNGIFNCRHCHNEAMNANEADPSKRHDLPRHKVERVICSLCGLEQDVHQVCSGCGVSMG 1747 CCN IF C HCH EA N D +RHD+PRH+VE+VIC LCG EQ+V Q+C CGV MG Sbjct: 76 CCNEIFGCHHCHYEAKNNINVDQKQRHDIPRHQVEQVICLLCGTEQEVGQICIHCGVCMG 135
Query: 1748 DYYCSICRFFDDDVSKGQFHCDSCGICRVGGQEKFFHCDKCGCCYAVALQKGHSCVENSM 1807 Y+C +C+ +DDD SK Q+HCD CGICR+GG+E FFHC KCGCCY++ L+ GH CVE +M Sbjct: 136 KYFCKVCKLYDDDTSKKQYHCDGCGICRIGGRENFFHCYKCGCCYSILLKNGHPCVEGAM 195
Query: 1808 HHNCPVCFDYLFDSTSDITVLRCGHTIHSECLREMTLHAQFSCPVCSKSVCDMSSAWERL 1867 HH+CP+CF++LF+S +D+TVL CGHTIH +CL EM H Q++CP+CSKSVCDMS WE+ Sbjct: 196 HHDCPICFEFLFESRNDVTVLPCGHTIHQKCLEEMRDHYQYACPLCSKSVCDMSKVWEKF 255
Query: 1868 DQEIAATPMPDAYRNKLV 1885 D EIAATPMP+ Y+N++V Sbjct: 256 DMEIAATPMPEPYQNRMV 273 Score = 310 bits (794), Expect = 7e-84, Method: Composition-based stats. Identities = 131/214 (61%), Positives = 156/214 (72%), Gaps = 9/214 (4%)
Query: 1266 KYGCPHYRRRCRIRAPCCNEVFGCRHCHNEAKGE-EADPRERHQIRRESIRRVICLLCDT 1324 +YGCPHYRRRC IRAPCCNE+FGC HCH EAK D ++RH I R + +VICLLC T Sbjct: 60 EYGCPHYRRRCCIRAPCCNEIFGCHHCHYEAKNNINVDQKQRHDIPRHQVEQVICLLCGT 119
Query: 1325 EQDVQQVCEGCGVCMGSYFCSKCNLFDDDTDKHQYHCDSCGICRVGGADNFFHCDRCGCC 1384 EQ+V Q+C CGVCMG YFC C L+DDDT K QYHCD CGICR+GG +NFFHC +CGCC Sbjct: 120 EQEVGQICIHCGVCMGKYFCKVCKLYDDDTSKKQYHCDGCGICRIGGRENFFHCYKCGCC 179
Query: 1385 YSVALQGKHVCVERAMHHNCPVCFEFLFDSVKQITVLQCGHTMHADCFNEMRLH------1438 YS+ L+ H CVE AMHH+CP+CFEFLF+S +TVL CGHT+H C EMR H Sbjct: 180 YSILLKNGHPCVEGAMHHDCPICFEFLFESRNDVTVLPCGHTIHQKCLEEMRDHYQYACP 239
Query: 1439 --SRSVLDLSEYWQTLDKEIAATPMPEALRGKTV 1470 S+SV D+S+ W+ D EIAATPMPE + + V Sbjct: 240 LCSKSVCDMSKVWEKFDMEIAATPMPEPYQNRMV 273
>ref|NP_566769.1| LAG1 (Longevity assurance gene 1) [Arabidopsis thaliana] Length = 310 /note="Identical to LAG1 longevity assurance homolog 1 (LAG1) [Arabidopsis Thaliana] (GB:Q9LDF2); similar to LAG13 (LAG1 LONGEVITY ASSURANCE HOMOLOG 3) [Arabidopsis thaliana] (TAIR:AT1G13580.2); similar to Lag1 longevity assurance-like 3 [Brassica rapa] (GB:ABV89617.1); contains InterPro domain TRAM, LAG1 and CLN8 homology; (InterPro:IPR006634); contains InterPro domain Longevity assurance proteins LAG1/LAC1; (InterPro:IPR016439); contains InterPro domain Longevity-assurance protein (LAG1); (InterPro:IPR005547)"
Score = 290 bits (742), Expect = 7e-78, Method: Composition-based stats. Identities = 144/292 (49%), Positives = 186/292 (63%), Gaps = 6/292 (2%)
Query: 898 REIDPSFWDLVTLAPIFAIGFPVCRFFLDRFVLEKLSRKSVFGTHESKLRKLSDADRDAL 957 +E P++ DL L P+FA+ FP RF LDRFV EKL+ ++G + + + DR Sbjct: 14 QESFPTYQDLGFL-PLFAVFFPTIRFLLDRFVFEKLASLVIYGRMSTN-KSDNIKDRKKN 71
Query: 958 RKTQTKFKESGWKCVYYTTAEIFALYVTYNETWLTDSYSIWVGPGDQTWPNQTIKVKLKL 1017 KFKES WKC+YY +AE+ AL VTYNE W +++ W+GPGDQ WP+Q +K+KLK Sbjct: 72 SPKVRKFKESAWKCIYYLSAELLALSVTYNEPWFSNTLYFWIGPGDQIWPDQPMKMKLKF 131
Query: 1018 LXXXXXXXXXXXXXXLIFWETRRKDFGVGSFNILVEPVKFVVLYFGASRFARIGCVVLAL 1077 L L+FWETRR DFGV + + V V+ Y R R G V+LAL Sbjct: 132 LYMFAAGFYTYSIFALVFWETRRSDFGVSMGHHITTLVLIVLSYI--CRLTRAGSVILAL 189
Query: 1078 HDASDVFLELAKMSKYAGVRVVPDVLFGLFALSWVLLRLIYFPVWVIWGTSYLSIKAINI 1137 HDASDVFLE+ KMSKY G + + F LFALSWV+LRLIY+P W++W TSY I ++ Sbjct: 190 HDASDVFLEIGKMSKYCGAESLASISFVLFALSWVVLRLIYYPFWILWSTSYQIIMTVDK 249
Query: 1138 HLHRGYGPIYYYVTNTLLISLFVLHIYWWVLIYRMIVKQIR-AGVIGDDVRS 1188 H GPI YY+ NTLL L VLHI+WWVLIYRM+VKQ++ G + +DVRS Sbjct: 250 EKHPN-GPILYYMFNTLLYFLLVLHIFWWVLIYRMLVKQVQDRGKLSEDVRS 300
Multalign to Arabidopsis Zinc-finger protein C3HC4-type RING finger The Blast results indicate that this gene prediction model is actually comprised of at least 3 different proteins, not just one. It seems as though it is mainly comprised of zinc-finger type proteins. This is one of the better Blast hits (using TAIR database). It has a pretty good alignment to the gene prediction model.
Multalign to Arabidopsis catalytic gene Multalign to Physcomitrella unknown protein These two Multalign alignments are fairly good. Both Physcomitrella and Arabidopsis have an exon (or extension of an exon) that is not present in the gene model prediction. Multalign Arabidopsis catalytic gene to Physcomitrella unknown protein
This alignment just shows that the two sequences that came up as strong Blast hits are very similar. Perhaps the Physcomitrella unknown protein has similar function as the Arabidopsis protein? Multalign to Arabidopsis longevity assurance protein
This alignment is also pretty good. There are numerous regions that seem to be conserved.
Based on the Blast results and Multalign alignments it seems as though this gene prediction model is comprised of 3 genes, a long gene with some catalytic function, a longevity assurance gene, and a zinc finger protein.