Supplement information:
Supplementary Table 1 Primers used. Name Sequence(5’ to 3’) rDNAup-6.14-F TCGATCCTAAGGGGTGGCATAAC XPR2t-gpd-R GACCCGTGCAGGACATCCTACTGCG GGACACGGGCATCTCACTTGCGTA GPD1p-xpr2t-F CATACGCAAGTGAGATGCCCGTGTCC CGCAGTAGGATGTCCTGCACGGGTCTTT GPD1p-DD253524-R GAGGCGCGACGCCCTGCGCGCCATTGTTGATGTGTGTTTAATTCAAGAATG DD253524-GPD1p-F CATTCTTGAATTAAACACACATCAACAATGGCGCGCAGGGCGTCGCGCCTC DD253524-lip2t-R GAAGTTGTAAAGAGTGATAAATAGCTCAAGCATTCTTGTACAGCGGGAGCG lip2t-DD253524-F CGCTCCCGCTGTACAAGAATGCTTGAGCTATTTATCACTCTTTACAACTTC rDNAd-6.14-R CTTCGGTATGATAGGAAGAGCC TEF1p-Pme1-F GTTTAAACAGAGACCGGGTTGGCGGCGTATTT TEF-ura3.1-HH-R CGAGCTTACTCGTTTCGTCCTCACGGACTCATCAGTTCCGGGAATGATTCTTATACTCAGAAGG ura3.1-HH-F AGGACGAAACGAGTAAGCTCGTCTTCCGGTTCCGGGTACACCGGTTTTAGAGCTAGAAATAGCAA Mig1t-Sal1-R TGTCTGTCGACAAACCCAAAAGGGCCGAAGGCTG pTEFin-Sal1-F TGGAAGATCCGGTCGACAGAGACCGGGTTGGCGGCGTATTTGTG URA3-F GGTGTGTTCTGTGGAGCATTCT △ku80-yz-up-R CCCGAGTCCGTTACCAACAAGA
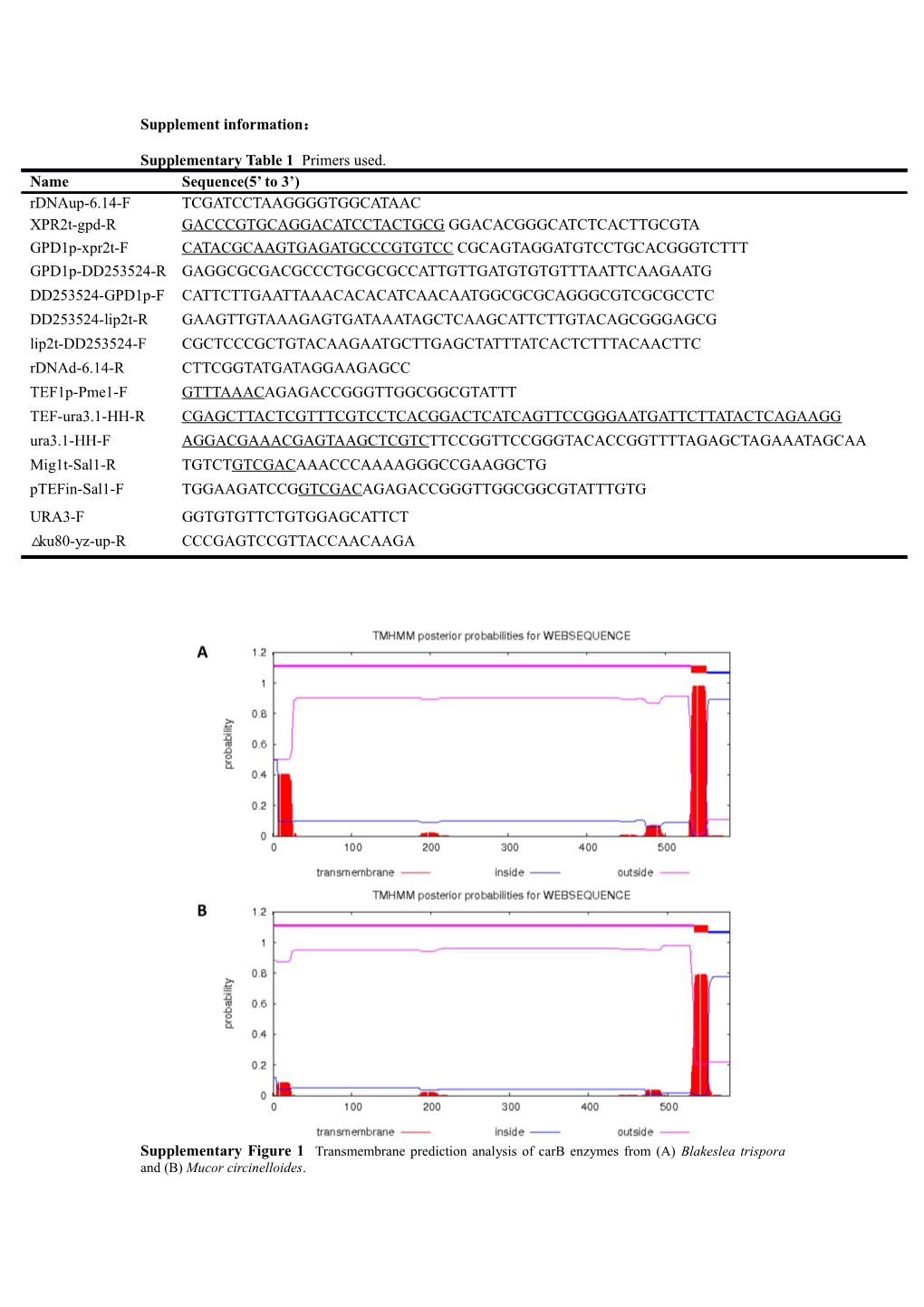
Supplementary Figure 1 Transmembrane prediction analysis of carB enzymes from (A) Blakeslea trispora and (B) Mucor circinelloides. Supplementary Figure 2 Transmembrane prediction analysis of (A) carRA enzyme from Blakeslea trispora and (B) carRP enzyme from Mucor circinelloides.
Supplementary Figure 3 URA3 disruption of CIBTS1961 via the CRISPR-Cas9 system. Transformants with the URA- phenotype appeared when CIBTS1961 was transformed with the plasmid pCAS1yl-ura3. Supplementary Figure 4 Disruption of URA3 in CIBTS1961, mediated by the CRISPR-Cas9 system. (A) Strategy for deletion of the URA3 gene from CIBTS1961. The URA3 gene was disrupted by insertion of donor DNA located on the same chromosome. The URA3 selection marker remained in chromosome E of the CIBTS1961 strain. The strains CIBTS1961 and MYA-2613 carry a ura3-302 mutation in their genomes. We found that there were two spliced regions, 3171155~3172275 bp and 3175134~3175553 bp, homologous to the URA3 cassette in chromosome E. It has been reported that competing alleles cut by CRISPR-Cas9 could be repaired by copying the endonuclease gene (DiCarlo et al., 2015). We tried to rescue the URA3 marker with the CRISPR-Cas9 system, in which these two spliced homologous sequences were used as donor DNA to disrupt the ura3 gene. The plasmid pCAS1yl-ura3, derived from pCen1-Cas9 carrying a Cas9 expression cassette and a ura3.sgRNA expression cassette, was constructed. 18 URA- strains were confirmed by colony PCR and sequencing. The results indicated that URA3 was successfully disrupted through homologous recombination with the donor DNA located in the same chromosome. (B) Confirmation of disruptants by colony PCR. Control, strain CIBTS1961. Lanes 1~18, transformants with the ura- phenotype.
Reference: DiCarlo, J. E., Chavez, A., Dietz, S. L., Esvelt, K. M., Church, G. M., 2015. Safeguarding CRISPR- Cas9 gene drives in yeast. Nat Biotech. 33, 1250-1255.