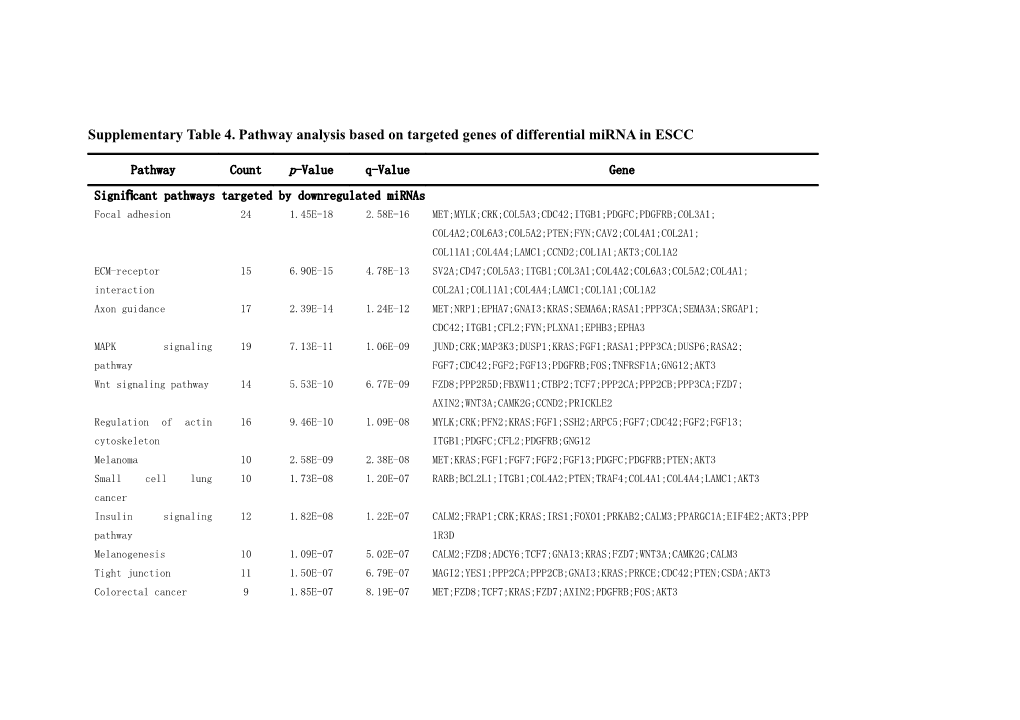
Supplementary Table 4. Pathway analysis based on targeted genes of differential miRNA in ESCC
Pathway Count p-Value q-Value Gene
Signifi cant pathways targeted by downregulated miRNAs Focal adhesion 24 1.45E-18 2.58E-16 MET;MYLK;CRK;COL5A3;CDC42;ITGB1;PDGFC;PDGFRB;COL3A1; COL4A2;COL6A3;COL5A2;PTEN;FYN;CAV2;COL4A1;COL2A1; COL11A1;COL4A4;LAMC1;CCND2;COL1A1;AKT3;COL1A2 ECM-receptor 15 6.90E-15 4.78E-13 SV2A;CD47;COL5A3;ITGB1;COL3A1;COL4A2;COL6A3;COL5A2;COL4A1; interaction COL2A1;COL11A1;COL4A4;LAMC1;COL1A1;COL1A2 Axon guidance 17 2.39E-14 1.24E-12 MET;NRP1;EPHA7;GNAI3;KRAS;SEMA6A;RASA1;PPP3CA;SEMA3A;SRGAP1; CDC42;ITGB1;CFL2;FYN;PLXNA1;EPHB3;EPHA3 MAPK signaling 19 7.13E-11 1.06E-09 JUND;CRK;MAP3K3;DUSP1;KRAS;FGF1;RASA1;PPP3CA;DUSP6;RASA2; pathway FGF7;CDC42;FGF2;FGF13;PDGFRB;FOS;TNFRSF1A;GNG12;AKT3 Wnt signaling pathway 14 5.53E-10 6.77E-09 FZD8;PPP2R5D;FBXW11;CTBP2;TCF7;PPP2CA;PPP2CB;PPP3CA;FZD7; AXIN2;WNT3A;CAMK2G;CCND2;PRICKLE2 Regulation of actin 16 9.46E-10 1.09E-08 MYLK;CRK;PFN2;KRAS;FGF1;SSH2;ARPC5;FGF7;CDC42;FGF2;FGF13; cytoskeleton ITGB1;PDGFC;CFL2;PDGFRB;GNG12 Melanoma 10 2.58E-09 2.38E-08 MET;KRAS;FGF1;FGF7;FGF2;FGF13;PDGFC;PDGFRB;PTEN;AKT3 Small cell lung 10 1.73E-08 1.20E-07 RARB;BCL2L1;ITGB1;COL4A2;PTEN;TRAF4;COL4A1;COL4A4;LAMC1;AKT3 cancer Insulin signaling 12 1.82E-08 1.22E-07 CALM2;FRAP1;CRK;KRAS;IRS1;FOXO1;PRKAB2;CALM3;PPARGC1A;EIF4E2;AKT3;PPP pathway 1R3D Melanogenesis 10 1.09E-07 5.02E-07 CALM2;FZD8;ADCY6;TCF7;GNAI3;KRAS;FZD7;WNT3A;CAMK2G;CALM3 Tight junction 11 1.50E-07 6.79E-07 MAGI2;YES1;PPP2CA;PPP2CB;GNAI3;KRAS;PRKCE;CDC42;PTEN;CSDA;AKT3 Colorectal cancer 9 1.85E-07 8.19E-07 MET;FZD8;TCF7;KRAS;FZD7;AXIN2;PDGFRB;FOS;AKT3 Prostate cancer 9 2.77E-07 1.15E-06 HSP90B1;FRAP1;TCF7;KRAS;FOXO1;PDGFC;PDGFRB;PTEN;AKT3 Glioma 8 3.03E-07 1.23E-06 CALM2;FRAP1;KRAS;PDGFRB;PTEN;CAMK2G;CALM3;AKT3 Gap junction 9 4.47E-07 1.69E-06 GJA1;ADCY6;GNAI3;KRAS;HTR2C;PDGFC;PDGFRB;TUBB2A;TUBB2B Calcium signaling 11 3.03E-06 9.27E-06 MYLK;CALM2;HTR2C;PTGFR;PPP3CA;PDGFRB;CAMK2G;CALM3;HTR7;ATP2B1;OXTR pathway Renal cell carcinoma 7 7.63E-06 1.96E-05 MET;CRK;KRAS;EGLN1;CDC42;ARNT;AKT3 GnRH signaling 8 1.35E-05 3.16E-05 CALM2;MAP3K3;ADCY6;KRAS;CDC42;CAMK2G;CALM3;HBEGF pathway Pathogenic 6 1.92E-05 4.23E-05 ARPC5;CDC42;ITGB1;TUBB2A;FYN;TUBB2B Escherichia coli infection - EHEC Pathogenic 6 1.92E-05 4.23E-05 ARPC5;CDC42;ITGB1;TUBB2A;FYN;TUBB2B Escherichia coli infection - EPEC Adipocytokine 6 5.97E-05 1.03E-04 FRAP1;IRS1;TNFRSF1A;PRKAB2;PPARGC1A;AKT3 signaling pathway Methionine metabolism 4 7.70E-05 1.26E-04 DNMT3A;MAT2A;DNMT3B;MAT1A T cell receptor 7 1.23E-04 1.90E-04 KRAS;PPP3CA;CDC42;FOS;FYN;IFNG;AKT3 signaling pathway Selenoamino acid 4 1.27E-04 1.92E-04 SEPHS2;GGT7;MAT2A;MAT1A metabolism Adherens junction 6 1.50E-04 2.23E-04 MET;YES1;TCF7;CDC42;PVRL3;FYN Endometrial cancer 5 1.78E-04 2.53E-04 TCF7;KRAS;AXIN2;PTEN;AKT3 Jak-STAT signaling 8 1.88E-04 2.62E-04 SPRED1;BCL2L1;IL6ST;GHR;SPRY1;CCND2;IFNG;AKT3 pathway Basal cell carcinoma 5 2.32E-04 3.14E-04 FZD8;TCF7;FZD7;AXIN2;WNT3A ErbB signaling 6 2.55E-04 3.43E-04 FRAP1;CRK;KRAS;CAMK2G;HBEGF;AKT3 pathway SNARE interactions in 4 5.70E-04 6.66E-04 SNAP25;VAMP2;STX5;STX1A vesicular transport p53 signaling pathway 5 6.69E-04 7.60E-04 SHISA5;SESN1;PTEN;CCND2;PPM1D Long-term 5 9.78E-04 0.001057 CALM2;KRAS;PPP3CA;CAMK2G;CALM3 potentiation Chronic myeloid 5 9.78E-04 0.001057 CRK;CTBP2;BCL2L1;KRAS;AKT3 leukemia Phosphatidylinositol 5 0.001038 0.001077 CALM2;DGKH;DGKD;PTEN;CALM3 signaling system Notch signaling 4 0.001283 0.001286 CTBP2;RBPJ;MAML1;NCOR2 pathway Glycerolipid 4 0.001283 0.001286 DGKH;LPL;DGKD;GPAM metabolism Cytokine-cytokine 9 0.001465 0.001437 MET;IL6ST;INHBB;GHR;PDGFC;PDGFRB;TNFRSF1A;IFNG;CX3CL1 receptor interaction TGF-beta signaling 5 0.001898 0.001762 GDF6;PPP2CA;PPP2CB;INHBB;IFNG pathway Alzheimer's disease 7 0.002213 0.002019 CALM2;COX6A1;PPP3CA;LPL;TNFRSF1A;ATP5G1;CALM3 Acute myeloid 4 0.002978 0.002625 FRAP1;TCF7;KRAS;AKT3 leukemia Aminosugars 3 0.003047 0.002663 GNPDA2;GFPT2;NANP metabolism Glycerophospholipid 4 0.005229 0.004164 PISD;DGKH;DGKD;GPAM metabolism Long-term depression 4 0.005786 0.004576 PPP2CA;PPP2CB;GNAI3;KRAS Pancreatic cancer 4 0.006381 0.005008 BCL2L1;KRAS;CDC42;AKT3 B cell receptor 4 0.007015 0.005485 KRAS;PPP3CA;FOS;AKT3 signaling pathway VEGF signaling 4 0.007347 0.005723 KRAS;PPP3CA;CDC42;AKT3 pathway Fc epsilon RI 4 0.008404 0.006334 KRAS;PRKCE;FYN;AKT3 signaling pathway Type II diabetes 3 0.01051 0.007684 FRAP1;PRKCE;IRS1 mellitus Apoptosis 4 0.012633 0.008907 BCL2L1;PPP3CA;TNFRSF1A;AKT3 Ubiquitin mediated 5 0.013544 0.009518 UBE2Q1;FBXW11;UBA2;UBA6;UBE2J1 proteolysis mTOR signaling 3 0.015548 0.010673 FRAP1;EIF4E2;AKT3 pathway Neuroactive ligand- 7 0.01561 0.010681 PTH1R;HTR2C;PTGFR;GHR;CNR1;HTR7;OXTR receptor interaction Non-small cell lung 3 0.017196 0.011538 RARB;KRAS;AKT3 cancer Amyotrophic lateral 3 0.018937 0.012505 ALDH5A1;GFPT2 sclerosis (ALS) Glutamate metabolism 2 0.024102 0.015215 BCL2L1;PPP3CA;TNFRSF1A Biosynthesis of 2 0.032153 0.01967 HMGCR;SQLE steroids Heparan sulfate 2 0.032153 0.01967 HS3ST3B1;HS3ST2 biosynthesis Epithelial cell 3 0.033765 0.020475 MET;CDC42;HBEGF signaling in Helicobacter pylori infection Thyroid cancer 2 0.034309 0.020715 TCF7;KRAS Folate biosynthesis 2 0.043466 0.025289 GCH1;ENTPD7 Regulation of 2 0.048346 0.027595 GABARAPL1;IFNG autophagy Parkinson's disease 4 0.049101 0.027595 COX6A1;UBE2J1;ATP5G1;GPR37 Natural killer cell 4 0.051299 0.028416 KRAS;PPP3CA;FYN;IFNG mediated cytotoxicity Basal transcription 2 0.053415 0.029431 TBPL1;TAF5 factors Butanoate metabolism 2 0.056017 0.030743 ALDH5A1;HMGCS1 Sphingolipid 2 0.058663 0.032023 SGPP1;SMPD3 metabolism ABC transporters - 2 0.072504 0.038373 ABCA1;ABCB6 General Proteasome 2 0.084249 0.042898 PSME4;IFNG Glutathione 2 0.090324 0.04538 GGT7;GPX7 metabolism Lysine degradation 2 0.093408 0.046816 SUV420H1;SUV420H2 Hedgehog signaling 2 0.112515 0.053371 FBXW11;WNT3A pathway Arachidonic acid 2 0.112515 0.053371 GGT7;GPX7 metabolism Leukocyte 3 0.123286 0.056733 GNAI3;CDC42;ITGB1 transendothelial migration PPAR signaling 2 0.15676 0.069081 ANGPTL4;LPL pathway Toll-like receptor 2 0.274059 0.109413 FOS;AKT3 signaling pathway Cell cycle 2 0.336967 0.130036 MCM2;CCND2 Cell adhesion 2 0.391086 0.148713 ITGB1;PVRL3 molecules (CAMs) Oxidative 2 0.408696 0.154472 COX6A1;ATP5G1 phosphorylation Systemic lupus 2 0.426054 0.159963 H3F3B;IFNG erythematosus Purine metabolism 2 0.453259 0.168655 ADCY6;IMPDH1 Olfactory 3 0.747233 0.2689 CALM2;CAMK2G;CALM3 transduction Significant pathways targeted by upregulated miRNAs Calcium signaling 6 1.06E-05 1.02E-04 ATP2B1;PLCB1;VDAC1;ATP2B2;ATP2B4;ADCY9 pathway Ubiquitin mediated 5 3.73E-05 2.47E-04 WWP1;UBE2A;SKP1;ITCH;UBE2G1 proteolysis Non-small cell lung 3 4.14E-04 0.001391 FOXO3;RAF1;RB1 cancer Inositol phosphate 3 4.37E-04 0.001404 INPP5B;PLCB1;PIK3C2A metabolism Acute myeloid 3 5.37E-04 0.001508 RPS6KB1;RAF1;PIM1 leukemia Phosphatidylinositol 3 0.001125 0.002358 INPP5B;PLCB1;PIK3C2A signaling system TGF-beta signaling 3 0.00166 0.00285 RPS6KB1;ACVR2A;SKP1 pathway Prostate cancer 3 0.001715 0.00285 HSP90B1;RAF1;RB1 Gap junction 3 0.002009 0.003093 PLCB1;RAF1;ADCY9 Melanogenesis 3 0.00276 0.003781 PLCB1;RAF1;ADCY9 GnRH signaling 3 0.00299 0.004013 PLCB1;RAF1;ADCY9 pathway Cell cycle 3 0.004031 0.004722 CDC25A;SKP1;RB1 Axon guidance 3 0.005157 0.005665 RASA1;SEMA4C;EPHA3 Bladder cancer 2 0.00565 0.006111 RAF1;RB1 Parkinson's disease 3 0.005843 0.006256 SNCA;VDAC1;UBE2G1 Insulin signaling 3 0.006083 0.006448 RPS6KB1;RAF1;IRS2 pathway Jak-STAT signaling 3 0.008363 0.008363 SPRY2;SPRED1;PIM1 pathway mTOR signaling 2 0.008552 0.008433 RPS6KB1;DDIT4 pathway Endometrial cancer 2 0.008552 0.008433 FOXO3;RAF1 Glioma 2 0.013117 0.011605 RAF1;RB1 Renal cell carcinoma 2 0.01551 0.012844 EPAS1;RAF1 Long-term depression 2 0.01551 0.012844 PLCB1;RAF1 Melanoma 2 0.01551 0.012844 RAF1;RB1 Pancreatic cancer 2 0.016346 0.013227 RAF1;RB1 Long-term 2 0.017201 0.013761 PLCB1;RAF1 potentiation Chronic myeloid 2 0.017201 0.013761 RAF1;RB1 leukemia Focal adhesion 3 0.01767 0.013977 RAF1;COL2A1;COL1A2 ECM-receptor 2 0.021277 0.015662 COL2A1;COL1A2 interaction ErbB signaling 2 0.022716 0.016492 RPS6KB1;RAF1 pathway Cell adhesion 2 0.049932 0.025818 MPZ;NLGN2 molecules (CAMs) Maturity onset 1 0.062017 0.029189 HHEX diabetes of the young Wnt signaling pathway 2 0.06237 0.029189 PLCB1;SKP1 Purine metabolism 2 0.06237 0.029189 POLE4;ADCY9 Heparan sulfate 1 0.071975 0.032397 HS3ST1 biosynthesis O-Glycan biosynthesis 1 0.079375 0.034696 OGT Alzheimer's disease 2 0.081154 0.035111 PLCB1;SNCA DNA polymerase 1 0.091579 0.038445 POLE4 Base excision repair 1 0.091579 0.038445 POLE4 Nucleotide excision 1 0.110772 0.044646 POLE4 repair Type II diabetes 1 0.113142 0.045172 IRS2 mellitus Regulation of actin 2 0.115052 0.045676 RAF1;PFN2 cytoskeleton Notch signaling 1 0.117865 0.046618 JAG1 pathway Proteasome 1 0.120217 0.047372 PSME3 Lysine degradation 1 0.127235 0.049676 SETD8 Basal cell carcinoma 1 0.136507 0.052809 GLI3 Hedgehog signaling 1 0.141106 0.05439 GLI3 pathway Cholera - Infection 1 0.145681 0.055748 ADCY9 Cytokine-cytokine 2 0.154599 0.058737 CCL1;ACVR2A receptor interaction Adipocytokine 1 0.16374 0.061617 IRS2 signaling pathway MAPK signaling 2 0.166741 0.062454 RASA1;RAF1 pathway p53 signaling pathway 1 0.168195 0.062777 CCNG2 B cell receptor 1 0.18142 0.067475 RAF1 signaling pathway VEGF signaling 1 0.183604 0.068049 RAF1 pathway Fc epsilon RI 1 0.19012 0.070219 RAF1 signaling pathway Colorectal cancer 1 0.200867 0.07342 RAF1 Small cell lung 1 0.205126 0.07472 RB1 cancer Antigen processing 1 0.211473 0.076505 PSME3 and presentation Pyrimidine metabolism 1 0.219856 0.079268 POLE4 T cell receptor 1 0.252516 0.086205 RAF1 signaling pathway Natural killer cell 1 0.308293 0.096115 RAF1 mediated cytotoxicity