Biol 470 Assignment (Due: Tuesday 4/15/08)
Estimating Evolutionary Trees Using DNA Sequence Data
Introduction
What is phylogenetics? It is the way that biologists reconstruct the pattern of events that have led to the distribution and diversity of life. There is an amazing diversity of life, both living and extinct. For biologists to communicate with each other about these many organisms, there must also be a classification of these organisms into groups. Ideally, the classification should be meaningful, and not arbitrary -- it should be based on the evolutionary history of life, such that it predicts properties of newly discovered or poorly known organisms.
Classification, however, is only one aspect of the much larger field of phylogenetic systematics. Systematics is an attempt to understand the evolutionary interrelationships of living things, trying to interpret the way in which life has diversified and changed over time. While classification is primarily the creation of names for groups, systematics goes beyond this to elucidate new theories of the mechanisms of evolution.
Systematics, then, is the study of the pattern of relationships among taxa; it is no less than understanding the history of all life. But history is not something we can see. It has happened once and leaves only clues as to the actual events. Biologists in general and systematists in particular use these clues to build hypotheses or models of the history.
Phylogenetic Trees: Presenting Evolutionary Relationships
Systematics describes the pattern of relationships among taxa and is intended to help us understand the history of all life. But history is not something we can see--it has happened once and leaves only clues as to the actual events. Scientists use these clues to build hypotheses, or models, of life's history. In phylogenetic studies, the most convenient way of visually presenting evolutionary relationships among a group of organisms is through illustrations called phylogenetic trees.
Some Definitions:
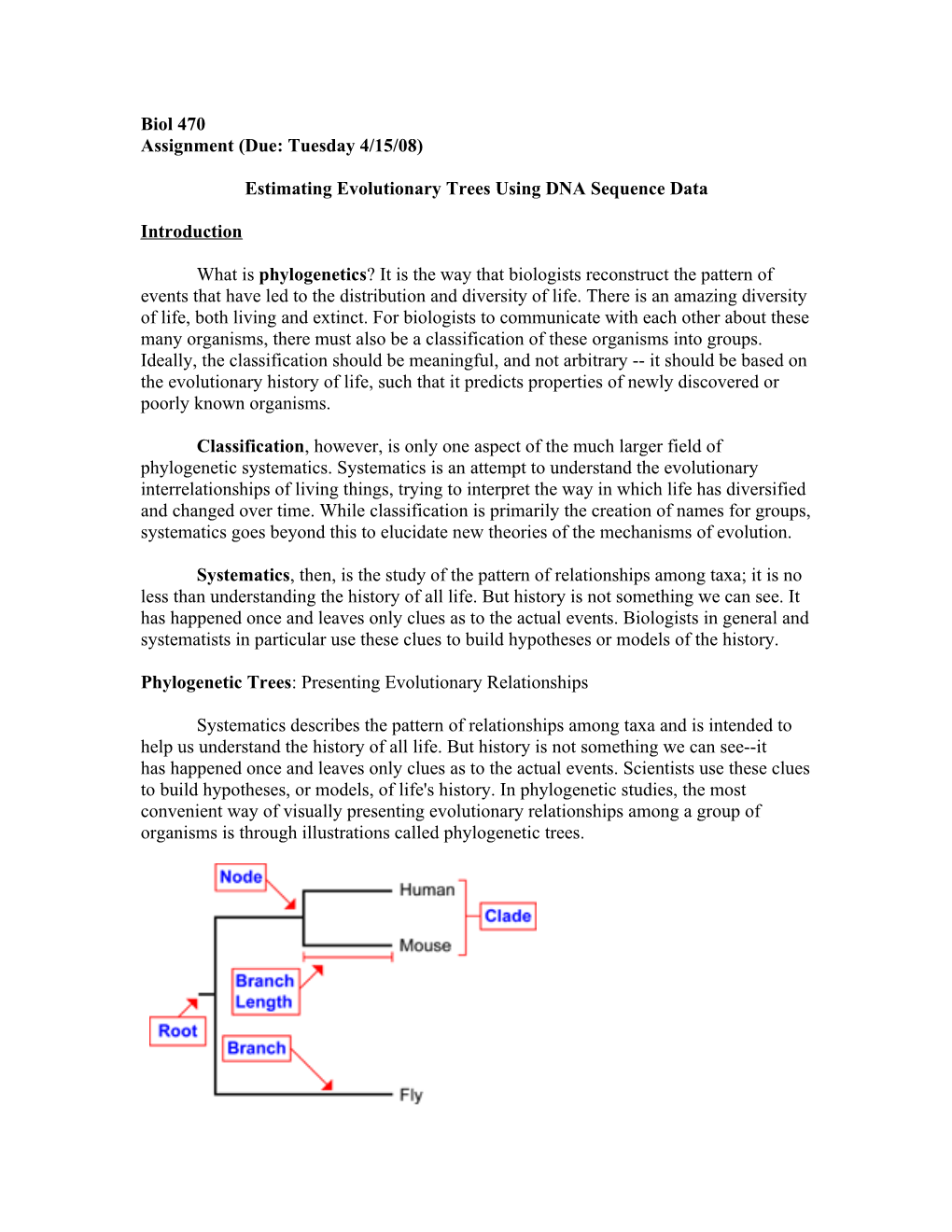
Node: represents a taxonomic unit. This can be either an existing species or an ancestor. Branch: Defines the relationship between the taxa in terms of descent and ancestry. Topology: The branching patterns of the tree. Branch length: Represents the number of character changes that have occurred in the branch. Root: The common ancestor of all taxa. Distance scale: scale that represents the number of differences between organisms or sequences. Clade: a group of two or more taxa or DNA sequences that includes both their common ancestor and all their descendents.
A phylogenetic tree is composed of nodes--each representing a taxonomic unit (species, populations, individuals)--and branches, which define the relationship between the taxonomic units in terms of descent and ancestry. Only one branch can connect any two adjacent nodes. The branching pattern of the tree is called the topology and the branch length usually represents the number of changes that have occurred in the branch. This is called a scaled branch. Scaled trees are often calibrated to represent the passage of time. Such trees have a theoretical basis in the particular gene or genes under analysis. Branches can also be unscaled, which means that the branch length is not proportional to the number of changes that has occurred, although the actual number may be indicated numerically somewhere on the branch. Phylogenetic trees may also be either rooted or unrooted. In rooted trees, there is a particular node, called the root--representing a common ancestor--from which a unique path leads to any other node. An unrooted tree only specifies the relationship among species, without identifying a common ancestor, or evolutionary path.
Molecular phylogenetics attempts to determine the rates and patterns of change occurring in DNA and proteins and to reconstruct the evolutionary history of genes and organisms. Two general approaches may be taken to obtain this information. In the first approach, scientists use DNA to study the evolution of an organism. In the second approach, different organisms are used to study the evolution of DNA. Whatever the approach, the general goal is to infer process from pattern: the processes of organismal evolution deduced from patterns of DNA variation and processes of molecular evolution inferred from the patterns of variations in the DNA itself. ASSIGNMENT: “Investigating Polar Bear and Giant Panda Ancestry”
Introduction
The animal order Carnivora contains four families: Canadiae (dogs, wolf), Felidae (small cats, large cats), Ursidae (bears), and Procyonidae (coatimundis, e.g., raccoons, lesser panda). The relationships of some carnivora, such as the giant panda and lesser panda, are a continuing controversy to taxonomists.
Classification of Bears (based on morphology) Kingdom Animalia Phylum Chordata Class Mammalia Order Carnivora Family Ursidae (bears) Genus Ailuropoda Ailuropoda melanoleuca (giant panda) Genus Ailurus Ailurus fulgens Genus Helarctos Helarctos malayanus (Malayan sun bear) Genus Melursus Melursus ursinus (sloth bear) Genus Tremarctos Tremarctos ornatus (spectacled bear) Genus Ursus Ursus americanus (American black bear) Ursus arctos (brown or grizzly bear) Ursus maritimus (polar bear) Ursus thibetanus (Asiatic black bear)
The giant panda is a specialist bamboo feeder and might well be the most popular wild animal worldwide. Is the giant panda a bear; or should it be classified in its own family? On the basis of comparative anatomical studies, karyological evidence, and palaeontological information, the giant panda has been classified into the Ursidae, the bear family. However, the giant panda shows differences from bears in genital structure, behavior, hemoglobin sequences, and restriction fragment length polymorphisms of mitochondrial DNA (mtDNA) that indicate that it is not closely related to bears.
Another outstanding question concerns the relationship between the polar bear and American brown bear. The geographic ranges of these two species are in close proximity. Did the polar bear evolve from a population of American brown bears that were isolated far north?
The genetic database (NCBI) www.ncbi.nlm.nih.gov contains DNA sequence data for a variety of organisms, including bears. To analyze the data you will use some of the applications of MEGA (Molecular Evolutionary Genetic Analysis) , a powerful sequence analysis software package available free of charge on the web. You will need to download this program.
Instructions:
You should work independently on this assignment.
(A) Go to the GenBank website: http://www.ncbi.nlm.nih.gov/Genbank/GenbankSearch.html
(B) Select “Entrez Browser” then “CoreNucleotide”. (C) Type in each accession number (see below for numbers; enter one at a time)
(D) Click on linked accession number
(E) what you will see is the genbank record for a portion of the mitochondrial 12S rRNA sequence for the American Black Bear. You can obtain all of the sequences using the accession numbers below, but I have already done this for you.
Species Accession Number American Black Bear Y08520 American Brown Bear (Grizzly) L21889 Spectacled Bear L21883 Asiatic Black Bear L21890 Polar Bear L22164 Giant Panda Y08521
(F) Go to my website www.csub.edu/~psmith3 and click on teaching. Then click on bear data. Save it to your desktop and rename the file as bear.meg
(G) click on bear.meg…this should launch MEGA right away.
(F) Note that the bear data has already been aligned. You could do this by eye or use MEGA. Below is the main page for MEGA. You could have aligned the data using the alignment option, you would probably need to download the free pdf manual for more thorough instructions. To use parsimony to analyze the bear data select “phylogeny” then “construct phylogeny” then “maximum parsimony” then “compute”. Just like that the analysis is finished. Select “image” from the tree menu that pops up and save as a tiff file. Next select “distances”, then “compute pairwise” then “compute”. This is a pairwise distance matrix. It indicates the proportion of nucleotides out of 350 total that differ between any two taxa being compared. Under “file” choose export/print distances.
When you launched MEGA a second window also will be shown see below. If you click on “C” it highlights all of the nucleotide columns that are identical. If you click on “V” it highlights all of the nucleotide columns that are variable. If you click on “Pi” it highlights all of the nucleotide columns that are parsimony informative (i.e., the synapomorphies). If you click on “S” it highlights the nucleotide columns where all of the bases are identical except 1 (i.e., apomorphies).
Your assignment: (1) copy the tree.tiff file into a word document. (2) Copy the pairwise distances into the word document…which two species were the most similar overall? What does this mean to you?. (3) out of a total of 350 bases how many (in %) were constant, variable, and parsimony informative? (4) Interpret the topology of the tree: specifically, in a paragraph answer the following questions: (1) Is the giant panda a bear? Explain. (2) Are the polar bear and American brown bear more closely related to each other than they are to other bears (indicating that one evolved from the other)? Explain. (3) Based on this tree the ancestor to the American Brown and Black bears probably originated from which geographic region?