Online Resource 1.
Genome wide gene expression profiles in response to plastid division perturbations. Planta. Jodi Maple, Per Winge, Astrid Elisabet Tveitaskog, Daniela Gargano, Atle M. Bones, Simon Geir Møller
Corresponding author: Simon Geir Møller, Centre for Organelle Research, Faculty of Science and Technology, University of Stavanger, Norway. [email protected]
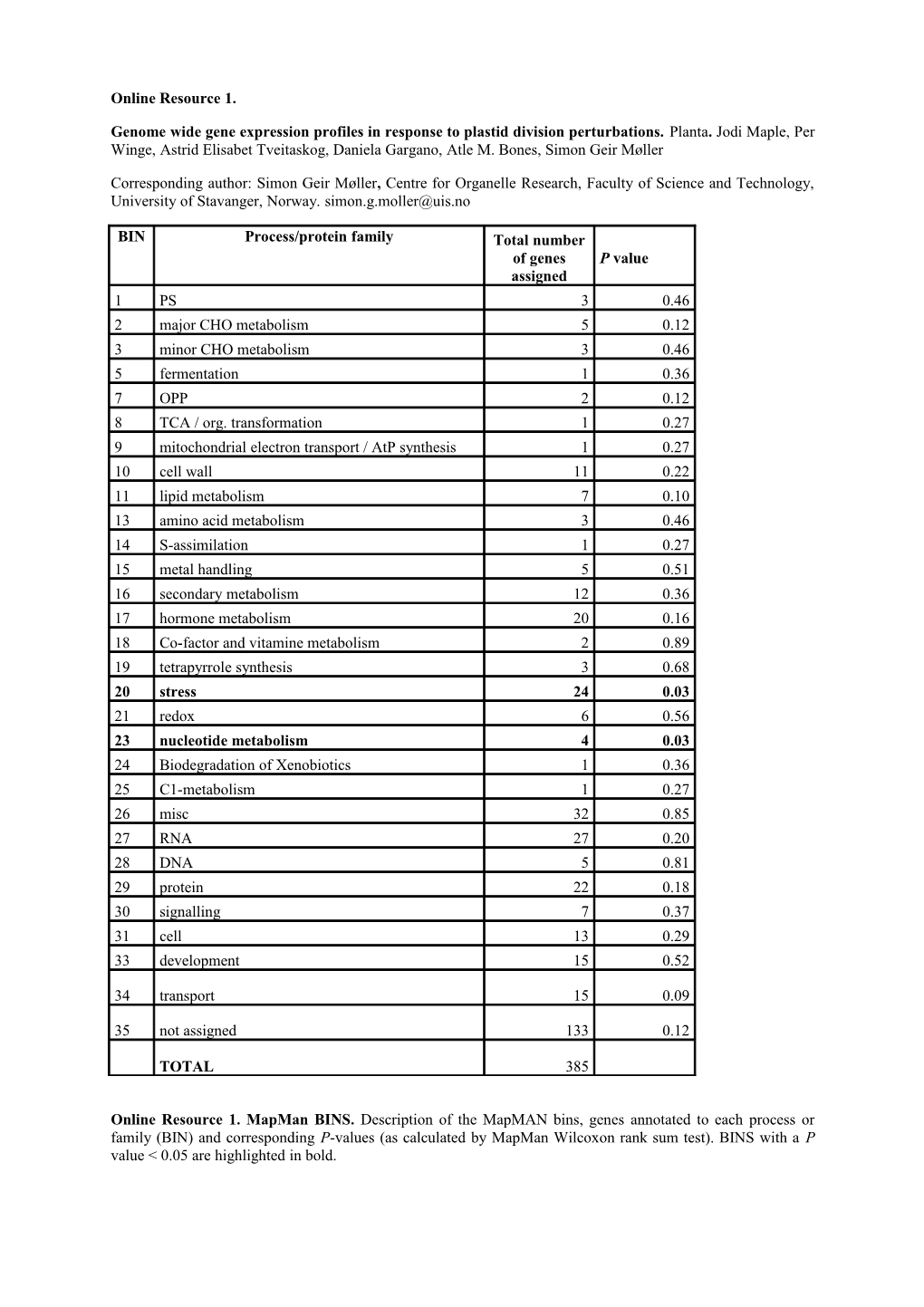
BIN Process/protein family Total number of genes P value assigned 1 PS 3 0.46 2 major CHO metabolism 5 0.12 3 minor CHO metabolism 3 0.46 5 fermentation 1 0.36 7 OPP 2 0.12 8 TCA / org. transformation 1 0.27 9 mitochondrial electron transport / AtP synthesis 1 0.27 10 cell wall 11 0.22 11 lipid metabolism 7 0.10 13 amino acid metabolism 3 0.46 14 S-assimilation 1 0.27 15 metal handling 5 0.51 16 secondary metabolism 12 0.36 17 hormone metabolism 20 0.16 18 Co-factor and vitamine metabolism 2 0.89 19 tetrapyrrole synthesis 3 0.68 20 stress 24 0.03 21 redox 6 0.56 23 nucleotide metabolism 4 0.03 24 Biodegradation of Xenobiotics 1 0.36 25 C1-metabolism 1 0.27 26 misc 32 0.85 27 RNA 27 0.20 28 DNA 5 0.81 29 protein 22 0.18 30 signalling 7 0.37 31 cell 13 0.29 33 development 15 0.52
34 transport 15 0.09
35 not assigned 133 0.12
TOTAL 385
Online Resource 1. MapMan BINS. Description of the MapMAN bins, genes annotated to each process or family (BIN) and corresponding P-values (as calculated by MapMan Wilcoxon rank sum test). BINS with a P value < 0.05 are highlighted in bold.