SUPPLEMENTARY DATA
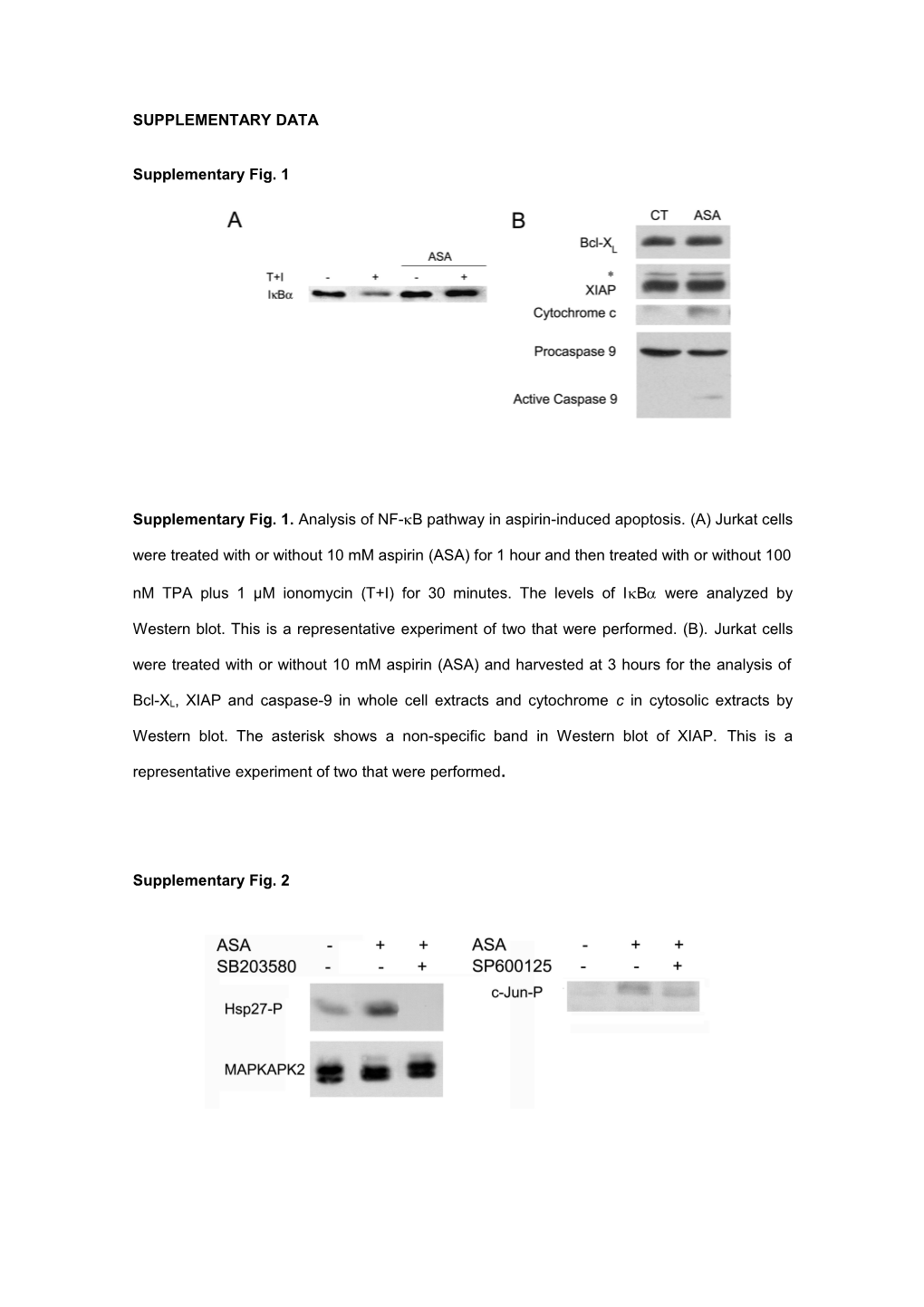
Supplementary Fig. 1
Supplementary Fig. 1. Analysis of NF-B pathway in aspirin-induced apoptosis. (A) Jurkat cells were treated with or without 10 mM aspirin (ASA) for 1 hour and then treated with or without 100 nM TPA plus 1 μM ionomycin (T+I) for 30 minutes. The levels of IB were analyzed by
Western blot. This is a representative experiment of two that were performed. (B). Jurkat cells were treated with or without 10 mM aspirin (ASA) and harvested at 3 hours for the analysis of
Bcl-XL, XIAP and caspase-9 in whole cell extracts and cytochrome c in cytosolic extracts by
Western blot. The asterisk shows a non-specific band in Western blot of XIAP. This is a representative experiment of two that were performed.
Supplementary Fig. 2 Supplementary Fig. 2. Measure of the activity of p38 MAPK and JNK induced by aspirin. Cells were pre-incubated with or without 10 M SB203580, p38 MAPK inhibitor (left panel) or 10 M
SP600125, JNK inhibitor (right panel) for 30 minutes, and then treated with 10 mM aspirin (ASA) for 15 minutes. p38 MAPK activity was assayed indirectly by immuno-precipitation of the downstream p38 MAPK kinase, the endogenous MAPKAPK 2 and its activity in immune complex kinase assays with Hsp27 as a substrate (Hsp27-P). An aliquot of the immunoprecipitate of each condition was analyzed by Western blot with the anti-MAPKAPK 2 antibody (left panels). The activity of JNK was analyzed by Western blot with a specific antibody against phosphorylated c-Jun (right panel). This is a representative experiment of two that were performed.
Supplementary Materials and Methods
MAPK-activated Protein Kinase 2 (MAPKAPK 2) activity
To assay the activity of MAPKAPK 2, protein extracts were obtained as described above for
MAPKs and 300 g subjected to immunoprecipitation with 1 g of anti-MAPKAPK 2 antibody from Santa Cruz Biotechnology (Santa Cruz, CA, USA) for 2.5 hours at 4ºC and protein G
Sepharose from Pharmacia Biotech (Cambridge, UK) for 1 h at 4ºC. An aliquot of the immune complex of each condition was assayed by Western blot with the anti-MAPKAPK 2 antibody from Santa Cruz Biotechnology (Santa Cruz, CA, USA), to confirm equal immunoprecipitation of
MAPKAPK 2. The activity of the immune complex was assayed at 30ºC for 30 minutes in 30 l
of kinase buffer (25 mM HEPES pH 7.5, 25 mM MgCl 2, 2 mM DTT, 100 M sodium orthovanadate, 25 mM -glycerophosphate) in the presence of 100 M ATP, 1Ci [32P] ATP and 1 g Hsp27 from StressGen Biotechnologies Corp. (Victoria, Canada) as a substrate.
Transient transfection and luciferase assay
Jurkat cells were transiently transfected by electroporation. For each electroporation, 15106 log- phase cells were collected, washed with RPMI and resuspended in 0.65 mL of complete medium without antibiotics. The cell suspension was mixed with 2 μg of the green fluorescent protein reporter plasmid pEGFP-C1 from Clontech (Palo Alto, CA, USA) and 10 μg of the luciferase reporter plasmid containing only the c-fos minimal promoter (the +52 /-110 region of the mouse c-fos gene, PGL-2fos) or the construct containing two consensus NF-B-binding sites (5´-
AGGGGACTTTCCGAGAGG-3´) in front of the c-fos minimal promoter (pNF-B-Luc) (kindly provided by F. Ventura and P. Muñoz, respectively). Cells were electroporated in 0.4 cm gap cuvettes at 250 V, 975 μF and 72 Ohms with the Electro cell manipulator 600 from BTX Inc.
(San Diego, CA, USA) and returned to complete medium. After 24 hours of incubation, transfection efficiency was determined by measuring the green fluorescence mean of each sample by flow cytometry (FACSCalibur) and the CellQuest software from Becton Dickinson
(Mountain View, CA, USA). At the same time, the remaining cells were treated with various factors as indicated and luciferase activity was assessed by lysing cells in Cell Culture Lysis
Reagent from Promega (Madison, WI, USA) (3106 cells in 130 μL) and mixing 40 μL of cellular lysates with 100 μL of Luciferase Assay Reagent from Promega (Madison, WI, USA). Light emission was recorded with the TD-20/20 Luminometer from Turner Designs (Sunnyvale, CA,
USA) at 10 seconds integration time. The protein concentration of each cell lysate was also determined with the Micro BCA Protein Assay Reagent kit from Pierce (Rockford, IL, USA).
Luciferase activity data correspond to luciferase activity standardised with the transfection efficiency and the protein concentration of each sample.
RT-MLPA
RNA was analysed by reverse transcriptase multiplex ligation-dependent probe amplification
(RT-MLPA), using SALSA MLPA KIT R011 Apoptosis mRNA from MRC-Holland (Amsterdam,
The Netherlands) (Eldering et al., Nucleic Acids Res 2003; 31:e153). In brief, RNA samples
(200 ng of total RNA) were first reverse-transcribed using a gene-specific probe mix. The resulting cDNA was annealed overnight at 60ºC to the MLPA probe mix. Annealed oligonucleotides were ligated by adding Ligase-65 and incubated at 54ºC for 15 minutes.
Ligation products were amplified by PCR (33 cycles, 30 seconds at 95ºC, 30 seconds at 60ºC and 1 minute at 72ºC) with one unlabelled and one FAM-labelled primer. The final PCR- amplified fragments were separated by capillary electrophoresis on a 48-capillary ABI-Prism
3730 Genetic Analyzer from Applied Biosystems/Hitachi (Foster City, CA, USA). The peak area and height were processed using GeneScan analysis software from Applied Biosystems (Foster
City, CA, USA). The levels of mRNA for each gene were expressed as a standardised ratio of the peak area divided by the peak area of a control gene. This resulted in the relative abundance of mRNAs of the genes of interest. The probe set contains probes for mRNAs of 30 apoptosis-related genes. Areas were standardised to -glucoronidase (GUS).