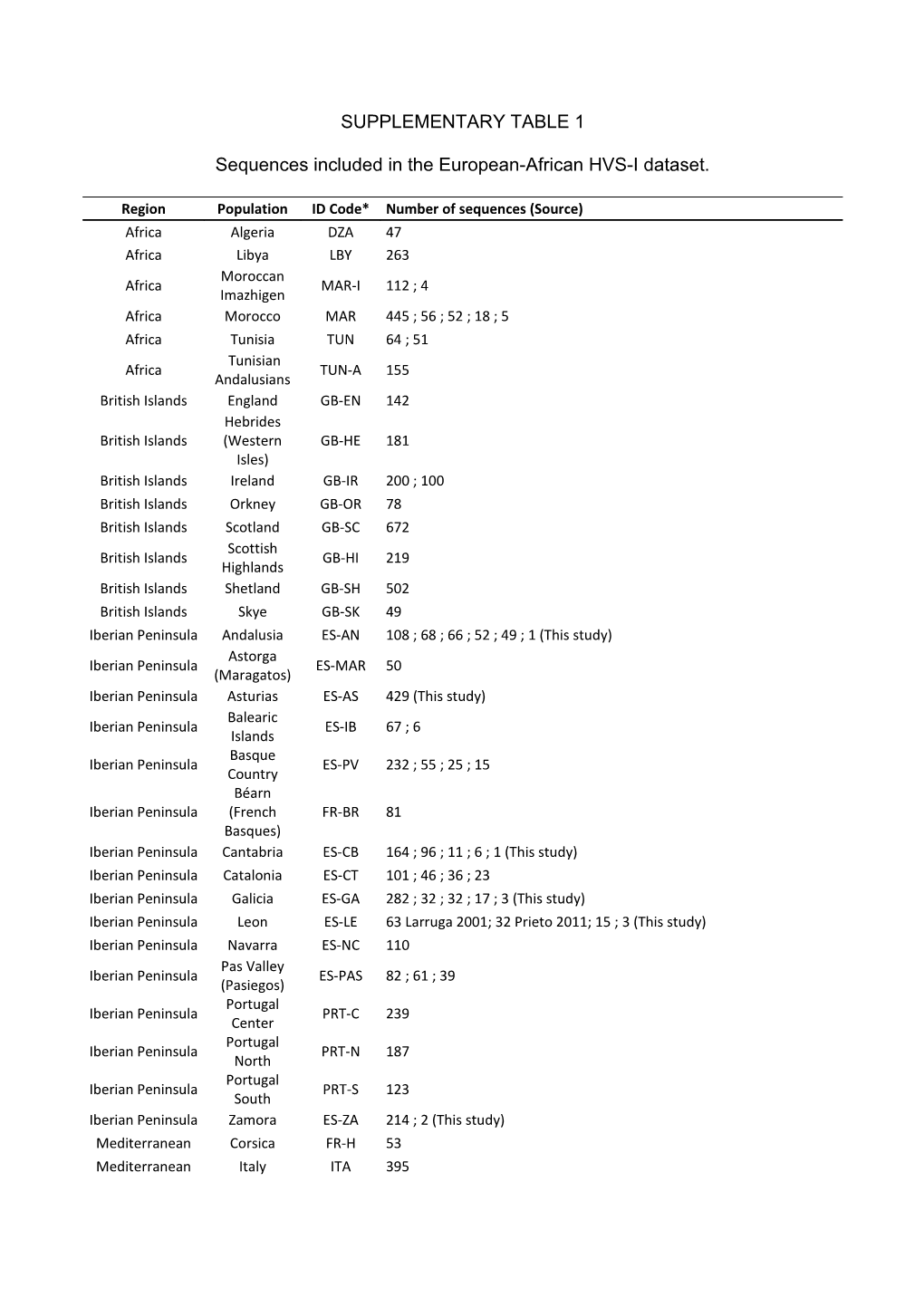
SUPPLEMENTARY TABLE 1
Sequences included in the European-African HVS-I dataset.
Region Population ID Code* Number of sequences (Source) Africa Algeria DZA 47 Africa Libya LBY 263 Moroccan Africa MAR-I 112 ; 4 Imazhigen Africa Morocco MAR 445 ; 56 ; 52 ; 18 ; 5 Africa Tunisia TUN 64 ; 51 Tunisian Africa TUN-A 155 Andalusians British Islands England GB-EN 142 Hebrides British Islands (Western GB-HE 181 Isles) British Islands Ireland GB-IR 200 ; 100 British Islands Orkney GB-OR 78 British Islands Scotland GB-SC 672 Scottish British Islands GB-HI 219 Highlands British Islands Shetland GB-SH 502 British Islands Skye GB-SK 49 Iberian Peninsula Andalusia ES-AN 108 ; 68 ; 66 ; 52 ; 49 ; 1 (This study) Astorga Iberian Peninsula ES-MAR 50 (Maragatos) Iberian Peninsula Asturias ES-AS 429 (This study) Balearic Iberian Peninsula ES-IB 67 ; 6 Islands Basque Iberian Peninsula ES-PV 232 ; 55 ; 25 ; 15 Country Béarn Iberian Peninsula (French FR-BR 81 Basques) Iberian Peninsula Cantabria ES-CB 164 ; 96 ; 11 ; 6 ; 1 (This study) Iberian Peninsula Catalonia ES-CT 101 ; 46 ; 36 ; 23 Iberian Peninsula Galicia ES-GA 282 ; 32 ; 32 ; 17 ; 3 (This study) Iberian Peninsula Leon ES-LE 63 Larruga 2001; 32 Prieto 2011; 15 ; 3 (This study) Iberian Peninsula Navarra ES-NC 110 Pas Valley Iberian Peninsula ES-PAS 82 ; 61 ; 39 (Pasiegos) Portugal Iberian Peninsula PRT-C 239 Center Portugal Iberian Peninsula PRT-N 187 North Portugal Iberian Peninsula PRT-S 123 South Iberian Peninsula Zamora ES-ZA 214 ; 2 (This study) Mediterranean Corsica FR-H 53 Mediterranean Italy ITA 395 Mediterranean Sardinia IT-SA 234 Mediterranean Sicily IT-SI 106 Mediterranean Tuscany IT-TO 322 ; 61 North Europe Iceland ISL 552 ; 394 North Europe Norway NOR 323 South Europe Brittany FR-E 236 South Europe France FRA 80 South Europe Languedoc FR-K 85 South Europe Lyonnais FR-V 46 South Europe Maine-Anjou FR-R 91 South Europe Normandie FR-P 46 South Europe Picardie FR-S 79 South Europe Poitou FR-T 124
* ID codes identify populations in the MDS plot (Figure 3), and are concordant with the ISO_3166-2 code of the location, when available. SUPPLEMENTARY TABLE 2
Asturian sequences and haplogroup assignments.
Coding Region Private to Sample Haplogroup HVS-I Polymorphisms (differences to rCRS) (RFLP) Asturias1 151 D4g1 16093C 16223T 16278T 16362C 16519C Yes 644 H* 16069T 16519C No 546 H* 16071G 16192T 16355T 7028C Yes 154 H* 16078C 16172C 16192T 16456A 16519C 7028C Yes 502 H* 16086C 16519C 7028C No 618 H* 16092C 16362C 16519C 7028C No 636 H* 16092C 16362C 16519C 7028C No 116 H* 16093C 16311C No 538 H* 16093C 16311C 16519C No 616 H* 16114T 16179T 16256T 16519C 7028C Yes 234 H* 16114T 16519C No 584 H* 16114T 16519C No 197 H* 16126C 16163G 16189C 16193.1CC Yes 624 H* 16129A 16298C 7028C No 244 H* 16140C 16519C No 8 H* 16148T 16304C 16390A 7028C No 591 H* 16166G 7028C No 182 H* 16169T 16519C 7028C No 346 H* 16169T 16519C 7028C No 153 H* 16172C 16192T 16456A 16519C 7028C No 342 H* 16176T 16519C No 603 H* 16183G 16265C 16293G 16311C 16327T 7028C Yes 580 H* 16188G 16519C 7028C No 136 H* 16188T 16519C 7028C No 194 H* 16189C 16193.1CC Yes 16189C 16194C 16258C 16274A 16293C 16297C 446 H* 7028C Yes 16298C 16434A 16475G 384 H* 16192T No 627 H* 16192T 16221T 16519C 7028C Yes 484 H* 16218T 16519C No 491 H* 16218T 16519C No 25 H* 16221T 16319A 16519C 7028C Yes 355 H* 16221T 16319A 16519C 7028C Yes 288 H* 16221T 16519C Yes 138 H* 16223T 16224C 16311C 16519C No 175 H* 16240G 16298C 7028C No 376 H* 16249C 16291T 16547T 16558A 7028C Yes 443 H* 16259T 16519C 7028C No 482 H* 16265T 16526A 7028C Yes 218 H* 16287T 16519C 7028C No 191 H* 16289G 16519C 7028C No 224 H* 16289G 16519C 7028C No 626 H* 16291T 16519C No 587 H* 16292T 16294T 16302G 16519C 7028C No 272 H* 16311C 16519C No 309 H* 16311C 16519C No 361 H* 16325C 16519C No 321 H* 16486G 16558A 16519C No 18 H* 16519C No 20 H* 16519C No 32 H* 16519C No 46 H* 16519C No 50 H* 16519C No 51 H* 16519C No 79 H* 16519C No 82 H* 16519C No 91 H* 16519C No 104 H* 16519C No 133 H* 16519C No 145 H* 16519C No 176 H* 16519C No 220 H* 16519C No 225 H* 16519C No 226 H* 16519C No 249 H* 16519C No 253 H* 16519C No 261 H* 16519C No 275 H* 16519C No 298 H* 16519C No 317 H* 16519C No 336 H* 16519C No 358 H* 16519C No 366 H* 16519C No 405 H* 16519C No 414 H* 16519C No 425 H* 16519C No 457 H* 16519C No 480 H* 16519C No 495 H* 16519C No 496 H* 16519C No 499 H* 16519C No 539 H* 16519C No 619 H* 16519C No 638 H* 16519C No 548 H* 16519C 16528T No 99 H* rCRS 7028C No 206 H* rCRS 7028C No 240 H* rCRS 7028C No 247 H* rCRS 7028C No 255 H* rCRS 7028C No 256 H* rCRS 7028C No 329 H* rCRS 7028C No 350 H* rCRS 7028C No 386 H* rCRS 7028C No 479 H* rCRS 7028C No 488 H* rCRS 7028C No 490 H* rCRS 7028C No 511 H* rCRS 7028C No 512 H* rCRS 7028C No 568 H* rCRS 7028C No 579 H* rCRS 7028C No 590 H* rCRS 7028C Yes 608 H* rCRS 7028C No 125 H1* 16032.1A 16105.1A 16185T 16519C 7028C 3010A Yes 521 H1* 16037G 16519C 7028C 3010A No 127 H1* 16092C 16362C 16519C 7028C 3010A No 600 H1* 16092C 16362C 16519C 7028C 3010A No 159 H1* 16093C 16519C 7028C 3010A No 273 H1* 16093C 16519C 7028C 3010A No 554 H1* 16093C 16519C 7028C 3010A No 556 H1* 16093C 16519C 7028C 3010A No 335 H1* 16114T 16519C 7028C 3010A No 262 H1* 16129A 16278T 16519C 7028C 3010A Yes 70 H1* 16129A 16519C 7028C 3010A No 284 H1* 16129A 16519C 7028C 3010A No 487 H1* 16140C 16519C 7028C 3010A No 463 H1* 16176T 16219G 16519C 7028C 3010A No 526 H1* 16176T 16219G 16519C 7028C 3010A No 123 H1* 16185T 16519C 7028C 3010A Yes 291 H1* 16189C 16519C 7028C 3010A No 325 H1* 16194C 16195A 7028C 3010A Yes 621 H1* 16222T 7028C 3010A Yes 164 H1* 16240G 16298C 7028C 3010A No 637 H1* 16278T 16519C 7028C 3010A No 486 H1* 16288C 16360T 16519C 7028C 3010A Yes 555 H1* 16304C 16362C 7028C 3010A Yes 202 H1* 16311C 16519C 7028C 3010A No 623 H1* 16357C 16519C 7028C 3010A No 13 H1* 16519C 7028C 3010A No 29 H1* 16519C 7028C 3010A No 35 H1* 16519C 7028C 3010A No 45 H1* 16519C 7028C 3010A No 122 H1* 16519C 7028C 3010A Yes 162 H1* 16519C 7028C 3010A No 166 H1* 16519C 7028C 3010A No 167 H1* 16519C 7028C 3010A No 198 H1* 16519C 7028C 3010A No 254 H1* 16519C 7028C 3010A No 257 H1* 16519C 7028C 3010A No 263 H1* 16519C 7028C 3010A No 282 H1* 16519C 7028C 3010A No 286 H1* 16519C 7028C 3010A No 287 H1* 16519C 7028C 3010A No 343 H1* 16519C 7028C 3010A No 431 H1* 16519C 7028C 3010A No 456 H1* 16519C 7028C 3010A No 459 H1* 16519C 7028C 3010A No 510 H1* 16519C 7028C 3010A No 529 H1* 16519C 7028C 3010A No 547 H1* 16519C 7028C 3010A No 563 H1* 16519C 7028C 3010A No 609 H1* 16519C 7028C 3010A No 652 H1* 16519C 7028C 3010A No 328 H1* rCRS 7028C 3010A No 458 H1* rCRS 7028C 3010A No 551 H1* rCRS 7028C 3010A No 274 H1a* 16162G 16519C 7028C 3010A No 596 H1a3 16051G 16067A 16162G 16519C 7028C 3010A Yes 216 H1a3 16051G 16162G 16218T 16519C 7028C 3010A Yes 628 H1a3 16051G 16162G 16465T 16519C 7028C 3010A No 246 H1a3 16051G 16162G 16519C 7028C 3010A No 337 H2a2b* 16291T 16519C No 570 H2a2b* 16291T 16519C No 165 H2a2b1 16235G 16291T No 178 H2a2b1 16235G 16291T No 193 H2a2b1 16235G 16291T No 195 H2a2b1 16235G 16291T No 265 H2a2b1 16235G 16291T No 278 H2a2b1 16235G 16291T No 283 H2a2b1 16235G 16291T No 324 H2a2b1 16235G 16291T No 558 H2a2b1 16235G 16291T Yes 651 H2a2b1 16235G 16291T No 404 H2a2b1 16235G 16291T 16519C No 24 H2b 16311C 16519C 16524C No 102 H3 16037G 16362C 16497G 16519C 7028C 6776C Yes 171 H3 16077G 7028C 6776C No 230 H3 16129A 16519C 7028C 6776C No 121 H3 16207G 16286T 16519C 7028C 6776C No 561 H3 16207G 16286T 16519C 7028C 6776C No 632 H3 16209C 16311C 16519C 7028C 6776C No 379 H3 16249C 16291T 7028C 6776C Yes 454 H3 16249C 16291T 7028C 6776C Yes 612 H3 16288C 16519C 7028C 6776C No 179 H3 16311C 16519C 7028C 6776C No 239 H3 16311C 16519C 7028C 6776C No 293 H3 16368C 16519C 7028C 6776C No 146 H3 16519C 7028C 6776C No 205 H3 16519C 7028C 6776C No 229 H3 16519C 7028C 6776C No 238 H3 16519C 7028C 6776C No 245 H3 16519C 7028C 6776C No 259 H3 16519C 7028C 6776C No 341 H3 16519C 7028C 6776C No 464 H3 16519C 7028C 6776C No 473 H3 16519C 7028C 6776C No 524 H3 16519C 7028C 6776C No 565 H3 16519C 7028C 6776C No 31 H3 rCRS 7028C 6776C No 251 H3 rCRS 7028C 6776C No 252 H3 rCRS 7028C 6776C No 320 H3 rCRS 7028C 6776C No 340 H3 rCRS 7028C 6776C No 545 H3 rCRS 7028C 6776C No 617 H3 rCRS 7028C 6776C No 531 H5 16192T 16294T 16304C 16320T 7028C Yes 533 H5 16192T 16294T 16304C 16320T 7028C Yes 543 H5 16266T 16274A 16294T 16304C 16519C 7028C No 300 H5 16304C 7028C Yes 267 H5 16304C 16325C 7028C No 613 H5 16304C 16519C No 647 H5 16304C 16519C No 264 H5 16304C 16568C No 139 H6* 16092C 16362C 16482G 7028C No 304 H6* 16249C 16324C 16362C 16482G 16519C 7028C Yes 331 H6* 16249C 16362C 16482G No 549 H6* 16249C 16362C 16482G No 109 H6* 16362C 16482G Yes 207 H6* 16362C 16482G No 517 H6* 16362C 16482G No 542 H6* 16362C 16482G No 585 H6* 16362C 16482G No 258 H6a1a1a 16311C 16362C 16482G No 301 H6a1a1a 16311C 16362C 16482G No 378 H7a1 16048A 16261T 16519C 7028C Yes 385 H7a1 16048A 16261T 16519C 7028C Yes 5 H9a 16093C 16168T 16519C 7028C Yes 57 HV* 16289G 16519C 7028T 13708A No 95 HV* rCRS 7028T 13708A No 592 HV0 16061C 16298C 7028T Yes 34 HV0 16129A 16298C 7028T No 360 HV0 16129A 16298C 7028T No 371 HV0 16129A 16298C 7028T No 315 HV0 16274A 16298C 7028T No 11 HV0 16298C 16519C 7028T No 115 HV0 16298C 16519C 7028T No 118 HV0 16298C 16519C 16524G 7028T No 639 HV4a* 16114T 16221T 16519C 7028T Yes 409 HV4a* 16221T 16519C 7028T Yes 469 HV4a* 16221T 16519C 7028T Yes 260 HV4a1a 16126C 16221T 16291T 7028T Yes 573 HV4a1a 16221T 16291T 7028T No 574 HV4a1a 16221T 16291T 7028T No 513 I* 16129A 16223T 16391A 16519C No 111 I1a 16129A 16172C 16223T 16311C 16391A 16519C No 451 I2a 16129A 16145A 16223T 16391A 16519C No 349 J* 16063C 16069T 16126C No 120 J* 16069T 16126C No 129 J* 16069T 16126C No 150 J* 16069T 16126C No 333 J* 16069T 16126C No 437 J* 16069T 16126C No 540 J* 16069T 16126C 16153A 16519C No 149 J* 16069T 16126C 16265G Yes 105 J* 16069T 16126C 16269G 16289G 16311C 7028T 13780A Yes 106 J* 16069T 16126C 16269G 16289G 16311C 7028T 13780A Yes 107 J* 16069T 16126C 16269G 16289G 16311C 7028T 13780A Yes 217 J* 16069T 16126C 16269G 16289G 16311C 7028T 13780A Yes 506 J* 16069T 16126C 16269G 16289G 16311C 7028T 13780A Yes 130 J* 16069T 16126C 16278T 16366T 16519C 7028T 13780A No 140 J* 16069T 16126C 16278T 16366T 16519C 7028T 13780A No 222 J* 16069T 16126C 16278T 16366T 16519C 7028T 13780A No 338 J* 16069T 16126C 16278T 16366T 16519C 7028T 13780A No 396 J* 16069T 16126C 16278T 16366T 16519C 7028T 13780A No 509 J* 16069T 16126C 16278T 16366T 16519C 7028T 13780A No 520 J* 16069T 16126C 16278T 16366T 16519C 7028T 13780A No 219 J* 16069T 16126C 16311C No 210 J* 16069T 16126C 16519C No 305 J* 16069T 16126C 16519C No 318 J* 16069T 16126C 16519C No 351 J* 16069T 16126C 16519C No 514 J* 16069T 16126C 16519C No 642 J* 16126C 16278T 16519C 7028T 13780A Yes 16069T 16126C 16145A 16153A 16172C 16222T 59 J1b1a1 Yes 16261T 16069T 16126C 16145A 16153A 16172C 16222T 92 J1b1a1 Yes 16261T 16069T 16126C 16145A 16153A 16172C 16222T 566 J1b1a1 Yes 16261T 16069T 16126C 16145A 16153A 16172C 16222T 9 J1b1a1 Yes 16261T 13708A 352 J1b1a1 16069T 16126C 16145A 16172C 16222T 16261T No 571 J1b1a1 16069T 16126C 16145A 16172C 16222T 16261T No 532 J1b1a1 16145A 16172C 16222T 16261T Yes 572 J1b1a1 16172C 16222T 16261T 7028T 13780A Yes 611 J2a1a 16069T 16126C 16145A 16231C 16261T No 67 J2b1a 16069T 16126C 16145A 16193T 16278T Yes 69 J2b1a 16069T 16126C 16145A 16193T 16278T Yes 97 J2b1a 16069T 16126C 16145A 16193T 16278T Yes 184 J2b1a 16069T 16126C 16145A 16193T 16278T Yes 201 J2b1a 16069T 16126C 16193T 16278T No 158 J2b1a 16069T 16126C 16193T 16278T 16287T No 248 J2b1a 16069T 16126C 16193T 16278T 16287T No 359 J2b1a 16069T 16126C 16193T 16278T 16519C No 327 J2b1a 16069T 16126C 16278T 16519C No 501 K* 16145A 16224C 16311C 16519C 7028T 9055A No 101 K* 16224C 16311C 16519C 7028T 9055A No 147 K* 16224C 16311C 16519C No 148 K* 16224C 16311C 16519C No 204 K* 16224C 16311C 16519C No 308 K* 16224C 16311C 16519C No 334 K* 16224C 16311C 16519C No 344 K* 16224C 16311C 16519C No 427 K* 16224C 16311C 16519C No 476 K* 16224C 16311C 16519C No 655 K* 16224C 16311C 16519C No 583 K1a1 16093C 16156A 16224C 16240C 16311C 16519C No 641 K1a1 16093C 16224C 16290T 16311C 16519C No 507 K1a11 16129A 16224C 16311C 16519C No 516 K1a5 16224C 16278T 16311C 16362C 16519C Yes 16126C 16129A 16224C 16311C 16319A 16463G 530 K1b1a Yes 16519C 183 K1b1a 16129A 16224C 16311C 16319A 16463G 16519C Yes 237 K1c2 16093C 16153A 16224C 16311C 16320T 16519C 7028T 9055A Yes 649 K2b1 16224C 16270T 16519C No 16126C 16187T 16189C 16223T 16264T 16270T 504 L1b No 16278T 16293G 16311C 500 L2a 16223T 16278T 16292T 16294T 16311C 16390A Yes 22 L3f1b4a 16209C 16223T 16311C 16355T 16519C Yes 227 L3f1b4a 16209C 16223T 16311C 16519C No 233 L3f1b4a 16209C 16223T 16311C 16519C No 347 L3f1b4a 16209C 16223T 16311C 16519C No 577 L3f1b4a 16209C 16223T 16311C 16519C No 16129A 16183del 16186T 16189C 16223T 243 M1* Yes 16249C 16311C 16129A 16183del 16186T 16189C 16223T 348 M1* Yes 16249C 16311C 16129A 16185T 16189del 16223T 16249C 553 M1b1 No 16311C 16519C 96 N1* rCRS 1719A 7028T No 445 N1b 16145A 16176G 16223T 16239T 16390A 16519C Yes 448 N1b 16145A 16176G 16223T 16239T 16390A 16519C Yes 128 R0a 16126C 16362C No 498 T* 16048A 16126C 16294T 16468C 16519C 7028T 13368G Yes 213 T* 16126C 16186T 16189C 16294T 16519C No 408 T* 16126C 16292T 16294T 16519C No 132 T* 16126C 16294T 16519C No 650 T* 16126C 16294T 16519C No 16126C 16163G 16186T 16189C 16294T 16491G 135 T1a* No 16519C 134 T1a* 16126C 16163G 16186T 16189C 16294T 16519C No 426 T1a* 16126C 16163G 16186T 16189C 16294T 16519C No 436 T1a* 16126C 16163G 16186T 16189C 16294T 16519C No 472 T1a* 16126C 16163G 16186T 16189C 16294T 16519C No 518 T1a* 16126C 16163G 16186T 16189C 16294T 16519C No 16126C 16163G 16186T 16189C 16294T 16519C 55 T1a* No 9055G 12308G 401 T1a* 16126C 16186T 16189C 16294T 16519C No 294 T1a* 16186T 16189del 16294T 16519C Yes 295 T1a* 16186T 16189del 16294T 16519C No 16126C 16163G 16186T 16189C 16261T 16294T 646 T1a2a No 16519C 228 T2* 16126C 16172C 16245T 16294T 16296T 16519C 7028T 13368G No 231 T2* 16126C 16209C 16294T 16296T 16519C Yes 593 T2b* 16061C 16126C 16294T 16296T 16304C 16519C Yes 645 T2b* 16093C 16294T 16296T 16304C 16519C 7028T 13368G Yes 203 T2b* 16126C 16129A 16294T 16296T 16304C 16519C No 16126C 16147T 16294T 16296T 16297C 16304C 161 T2b* No 16519C 170 T2b* 16126C 16294T 16296T 16304C 16519C No 209 T2b* 16126C 16294T 16296T 16304C 16519C No 223 T2b* 16126C 16294T 16296T 16304C 16519C No 299 T2b* 16126C 16294T 16296T 16304C 16519C No 326 T2b* 16126C 16294T 16296T 16304C 16519C No 462 T2b* 16126C 16294T 16296T 16304C 16519C No 560 T2b* 16126C 16294T 16296T 16304C 16519C No 575 T2b* 16126C 16294T 16296T 16304C 16519C No 615 T2b* 16126C 16294T 16296T 16304C 16519C No 625 T2b* 16126C 16294T 16296T 16304C 16519C No 656 T2b* 16126C 16294T 16296T 16304C 16519C No 190 T2b* 16126C 16294T 16296T 16304C 16519C 16524G No 489 T2b* 16126C 16294T 16296T 16304C 16519C 16566A No 528 T2b* 16126C 16294T 16304C 16519C No 610 T2b* 16126C 16294T 16304C 16519C No 598 T2b* 16294T 16304C 16362C 16519C Yes 345 T2b3a 16126C 16292T 16294T 16296T 16304C 16519C No 144 T2c 16126C 16292T 16294T 16519C No 527 T2c 16126C 16292T 16294T 16519C No 648 T2c 16126C 16292T 16294T 16519C No 268 T2e* 16126C 16153A 16294T 16296T 16519C No 630 T2e1 16061C 16126C 16153A 16209C 16294T 16519C Yes 142 T2e1 16126C 16153A 16209C 16294T 16519C No 557 T2e1 16126C 16153A 16209C 16294T 16519C No 604 T2e1 16126C 16153A 16209C 16294T 16519C No 211 T2e1 16126C 16153A 16294T 16519C No 72 U* rCRS 7028T 12308G No 212 U* rCRS 7028T 12308G No 466 U1a2 16166del 16183C 16189C 16249C 16311C No 468 U4* 16100G 16356C 16519C Yes 7028T 9055G 266 U4* 16129A 16356C 16519C No 12368G 470 U4* 16356C 16519C No 544 U4* 16356C 16519C No 90 U4a1 16134T 16356C 16519C No 112 U4a1 16134T 16356C 16519C No 113 U4a1 16134T 16356C 16519C No 114 U4a1 16134T 16356C 16519C No 126 U4a1 16134T 16356C 16519C Yes 633 U4a1 16134T 16356C 16519C No 7028T 9055G 307 U4a3 16265G 16356C 16362C 16519C No 12368G 65 U5* 16192T 16270T 16274A Yes 58 U5* 16192T 16270T 16274A 16519C Yes 94 U5* 16192T 16270T 16274A 16519C Yes 310 U5* 16192T 16270T 16319A No 41 U5a1* 16169T 16192T 16256T 16270T 16399G Yes 42 U5a1* 16169T 16192T 16256T 16270T 16399G Yes 296 U5a1* 16169T 16192T 16256T 16270T 16399G Yes 508 U5a1* 16169T 16192T 16256T 16270T 16399G Yes 332 U5a1* 16192T 16256T 16270T 16294T 16399G Yes 12 U5a1* 16192T 16256T 16270T 16399G No 525 U5a1* 16256T 16270T 16399G No 75 U5a1b1* 16192T 16256T 16270T 16291T 16399G No 494 U5a1b1e 16270T 16291T 16294T 16399G Yes 614 U5a2 16172C 16192T 16256T 16270T 16526A Yes 492 U5a2 16192T 16256T 16270T 16526A No 16074G 16183C 16189C 16192T 16270T 16278T 7028T 9055G 323 U5b1* Yes 16291T 16384A 16434A 12368G 552 U5b1* 16189C 16192T 16270T No 595 U5b1* 16189C 16192T 16270T No 250 U5b1d 16224C 16270T 16519C No 7028T 9055G 277 U5b1d 16261T 16270T 16390A No 12368G 199 U5b1d 16270T 16519C No 200 U5b1d 16270T 16519C No 26 U5b3 16192T 16270T 16304C 16362C Yes 7028T 9055G 141 U6 16172C 16174T 16219G 16311C 16335G Yes 12368G 7028T 9055G 312 U6 16172C 16174T 16219G 16311C 16335G Yes 12368G 174 U6 16172C 16183C 16189C 16384A 16386A 16519G Yes 503 U6 16172C 16219G 16311C 16519C No 285 U6 16172C 16519C Yes 98 U8b 16129A 16519C 7028T 9055A No 19 U8b 16140C 16519C 7028T 9055A No 68 U8b rCRS 7028T 9055A No 71 U8b rCRS 7028T 9055A No 567 V 16075C 16240G 16298C 4580A 7028T Yes 241 V 16192T 16298C 4580A 7028T No 163 V 16240G 16298C 4580A 7028T No 177 V 16240G 16298C 4580A 7028T No 523 V 16274A 16298C 4580A 7028T No 7 V 16298C 4580A 7028T No 497 V 16298C 4580A 7028T No 137 V 16298C 16519C 4580A 7028T No 270 V 16298C 16519C 4580A 7028T No 242 W 16223T 16292T 16519C No 478 W 16223T 16292T 16519C No 519 W 16223T 16292T 16519C No
1 Relative to our HVS-I dataset of 9,539 additional European and North African sequences. SUPPLEMENTARY TABLE 3
Equivalences between populations used to calculate diversity values by Garcia et al. (2011) and populations from our HVS-I dataset used to calculate θK and the proportion of private lineages.
Population code Garcia et al. (2011) N This study N Galicia, Asturias, Pasiegos, Potes, Cantabria, Basques Galicia, Asturias, from Vizcaya, Basques IPNW 995 Cantabria, Basque 1507 from Guipuzcoa, Basques Country, Navarra from Alava, Basques Miscelanea Aragon, Catalonia, IPNE 438 Catalonia, Balearic Islands 279 Valencia Extremadura, Murcia, Andalucia, Leon, Leon, Zamora, Andalucia, IPS 1484 1222 Maragatos, Castilla, Spain Portugal Miscelanea, Portugal Béarn, Languedoc, Béarn, Languedoc, Provence, Limousin, Provence, Lyonnais, FRA Lyonnais, Poitou, Brittany, 1204 Poitou, Brittany, 551 Normandie, Picardie, Normandie, Picardie, France Miscelanea France Miscelanea Ireland, Scotland, Scottish Ireland, Scotland, Highlands, England, WIS England, Wales, Cornwall, 1610 2143 Hebrides, Orkney, Great Britain Miscelanea Shetland, Skye Germany, Austria, Switzerland, Poland, Germany*, Austria*, NCE 1802 714* North-Central Europe Switzerland* Miscelanea Corsica, Italy, Sicily, Macedonia, Greece, Corsica, Italy, Sardinia, MED 2282 1171 Croatia, Albania, South- Sicily, Tuscany East Europe Miscelanea Iceland, Scandinavia, SCA 849 Iceland, Norway 1269 Finland, Estonia, Karelians NER Russia, Volga 235 European Russia* 215* Tunez, Morocco, Argelia, Tunez, Morocco, Argelia, NAF Sahara, Mauritanians, 1065 Imazhigen, Andalusians, 1272 Imazhigen, Egypt Libya
* θK and private lineages values are data from Helgason et al. (2001). SUPPLEMENTARY TABLE 4
Description of the models tested in MIGRATE and Bayes factor computations.
Model Panmixia* Migration Likelihood Log (Bayes Factor) Prob (Bezier) Full No. Full matrix. -5731.488333 -54.581538 1.4053E-12 Restricted to 1 No. -6008.871272 -609.347416 4.8072E-133 adjacent. Basque populations are Restricted to 2 panmictic. Asturias and -5704.197564 0 ≈1 adjacent. Cantabria are panmictic. The Franco-Cantabrian area Restricted to 3 is panmictic. -5937.32334 -466.251552 5.6854E-102 adjacent. Asturias is excluded. The Franco-Cantabrian area Restricted to 4 is panmictic. -6231.796188 -1055.19725 7.3592E-230 adjacent. Asturias is included.
* French populations were considered a panmictic unit in all models. Additional calculations with varying degrees of panmixia did not alter the final model choice. SUPPLEMENTARY REFERENCES
Achilli A, Olivieri A, Pala M, Metspalu E, Fornarino S, Battaglia V, Accetturo M, Kutuev I, Khusnutdinova E, Pennarun E et al. . 2007. Mitochondrial DNA Variation of Modern Tuscans Supports the Near Eastern Origin of Etruscans. Am J Hum Genet 80:759-768.
Alfonso-Sánchez MA, Cardoso S, Martínez-Bouzas C, Peña JA, Herrera RJ, Castro A, Fernández-Fernández I, and De Pancorbo MM. 2008. Mitochondrial DNA haplogroup diversity in Basques: A reassessment based on HVI and HVII polymorphisms. Am J Hum Biol 20(2):154- 164.
Álvarez-Iglesias V, Mosquera-Miguel A, Cerezo M, Quintáns B, Zarrabeitia MT, Cuscó I, Lareu MV, García O, Pérez-Jurado L, and Carracedo A. 2009. New population and phylogenetic features of the internal variation within mitochondrial DNA macro-haplogroup R0. PLoS ONE 4(4):e5112.
Alvarez JC, Johnson DLE, Lorente JA, Martinez-Espin E, Martinez-Gonzalez LJ, Allard M, Wilson MR, and Budowle B. 2007. Characterization of human control region sequences for Spanish individuals in a forensic mtDNA data set. Leg Med 9(6):293-304.
Alvarez L, Santos C, Ramos A, Pratdesaba R, Francalacci P, and Aluja MP. 2010. Mitochondrial DNA patterns in the Iberian Northern plateau: population dynamics and substructure of the Zamora province. Am J Phys Anthropol 142(4):531-539.
Cali F, Le Roux M, D’Anna R, Flugy A, De Leo G, Chiavetta V, Ayala G, and Romano V. 2001. MtDNA control region and RFLP data for Sicily and France. Int J Legal Med 114(4):229-231.
Cardoso S, Alfonso-Sánchez MA, Valverde L, Odriozola A, Pérez-Miranda AM, Peña JA, and de Pancorbo MM. 2011. The maternal legacy of Basques in northern navarre: New insights into the mitochondrial DNA diversity of the Franco-Cantabrian area. Am J Phys Anthropol (In Press).
Cardoso S, Zarrabeitia MT, Valverde L, Odriozola A, Alfonso-Sanchez MA, and de Pancorbo MM. 2010. Variability of the entire mitochondrial DNA control region in a human isolate from the Pas Valley (northern Spain). J Forensic Sci 55(5):1196-1201.
Casas MJ, Hagelberg E, Fregel R, Larruga JM, and González AM. 2006. Human mitochondrial DNA diversity in an archaeological site in al Andalus: Genetic impact of migrations from North Africa in medieval Spain. Am J Phys Anthropol 131(4):539-551.
Cherni L, Fernandes V, Pereira JB, Costa MD, Goios A, Frigi S, Yacoubi-Loueslati B, Amor MB, Slama A, Amorim A et al. . 2009. Post-last glacial maximum expansion from Iberia to North Africa revealed by fine characterization of mtDNA H haplogroup in Tunisia. Am J Phys Anthropol 139(2):253-260.
Fadhlaoui-Zid K, Rodríguez-Botigué L, Naoui N, Benammar-Elgaaied A, Calafell F, and Comas D. 2011. Mitochondrial DNA structure in North Africa reveals a genetic discontinuity in the Nile Valley. Am J Phys Anthropol 145(1):107-117.
Falchi A, Giovannoni L, Calo CM, Piras IS, Moral P, Paoli G, Vona G, and Varesi L. 2005. Genetic history of some western Mediterranean human isolates through mtDNA HVR1 polymorphisms. J Hum Genet 51(1):9-14.
Garcia O, Fregel R, Larruga JM, Alvarez V, Yurrebaso I, Cabrera VM, and Gonzalez AM. 2011. Using mitochondrial DNA to test the hypothesis of a European post-glacial human recolonization from the Franco-Cantabrian refuge. Heredity 106(1):37-45. Goodacre S, Helgason A, Nicholson J, Southam L, Ferguson L, Hickey E, Vega E, Stefansson K, Ward R, and Sykes B. 2005. Genetic evidence for a family-based Scandinavian settlement of Shetland and Orkney during the Viking periods. Heredity 95(2):129-135.
Helgason A. 2000. mtDNA and the origin of the Icelanders: deciphering signals of recent population history. Am J Hum Genet 66(3):999-1016.
Helgason A, Hickey E, Goodacre S, Bosnes V, Stefánsson K, Ward R, and Sykes B. 2001. mtDNA and the islands of the North Atlantic: estimating the proportions of Norse and Gaelic ancestry. Am J Hum Genet 68(3):723-737.
Helgason A, Hrafnkelsson B, Gulcher JR, Ward R, and Stefánsson K. 2003. A populationwide coalescent analysis of Icelandic matrilineal and patrilineal genealogies: evidence for a faster evolutionary rate of mtDNA lineages than Y chromosomes. Am J Hum Genet 72(6):1370-1388.
Larruga JM, Díez F, Pinto FM, Flores C, and González AM. 2001. Mitochondrial DNA characterisation of European isolates: the Maragatos from Spain. Eur J Hum Genet 9(9):708.
Maca-Meyer N, Sánchez-Velasco P, Flores C, Larruga J-M, González A-M, Oterino A, and Leyva-Cobián F. 2003. Y Chromosome and Mitochondrial DNA Characterization of Pasiegos, a Human Isolate from Cantabria (Spain). Ann Hum Genet 67:329-339.
McEvoy B, Richards M, Forster P, and Bradley DG. 2004. The Longue Duree of genetic ancestry: multiple genetic marker systems and Celtic origins on the Atlantic facade of Europe. Am J Hum Genet 75(4):693-702.
Pereira L, Cunha C, and Amorim A. 2004. Predicting sampling saturation of mtDNA haplotypes: an application to an enlarged Portuguese database. Int J Legal Med 118(3):132-136.
Plaza S, Calafell F, Helal A, Bouzerna N, Lefranc G, Bertranpetit J, and Comas D. 2003. Joining the pillars of Hercules: mtDNA sequences show multidirectional gene flow in the western Mediterranean. Ann Hum Genet 67(4):312-328.
Prieto L, Zimmermann B, Goios A, Rodriguez-Monge A, Paneto GG, Alves C, Alonso A, Fridman C, Cardoso S, Lima G et al. . 2011. The GHEP-EMPOP collaboration on mtDNA population data--A new resource for forensic casework. Forensic Sci Int Genet (In Press, Corrected Proof).
Rhouda T, Martínez-Redondo D, Gómez-Durán A, Elmtili N, Idaomar M, and Díez-Sánchez C. 2009. Moroccan mitochondrial genetic background suggests prehistoric human migrations across the Gibraltar Strait. Mitochondrion 9(6):402-407.
Richard C, Pennarun E, Kivisild T, Tambets K, Tolk H-V, Metspalu E, Reidla M, Chevalier S, Giraudet S, Lauc LB et al. . 2007. An mtDNA perspective of French genetic variation. Ann Hum Biol 34(1):68-79.
Richards M, Corte-Real H, Forster P, Macaulay V, Wilkinson-Herbots H, Demaine A, Papiha S, Hedges R, Bandelt H-J, and Sykes B. 1996. Paleolithic and neolithic lineages in the European mitochondrial gene pool. Am J Hum Genet 59(1):185.
Turchi C, Buscemi L, Giacchino E, Onofri V, Fendt L, Parson W, and Tagliabracci A. 2009. Polymorphisms of mtDNA control region in Tunisian and Moroccan populations: an enrichment of forensic mtDNA databases with Northern Africa data. Forensic Sci Int Genet 3(3):166-172.
Turchi C, Buscemi L, Previderè C, Grignani P, Brandstätter A, Achilli A, Parson W, and Tagliabracci A. 2008. Italian mitochondrial DNA database: results of a collaborative exercise and proficiency testing. Int J Legal Med 122(3):199-204.