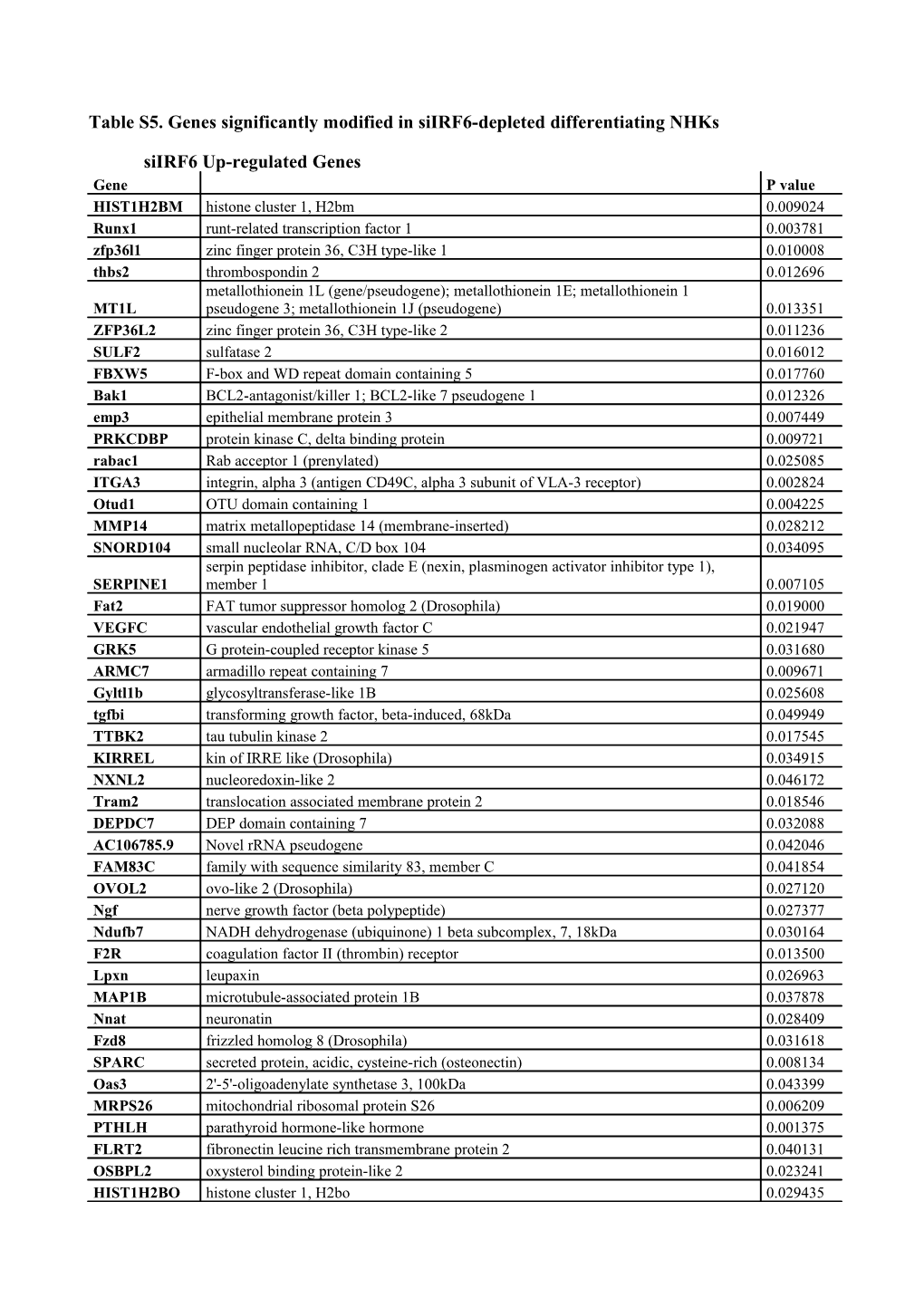
Table S5. Genes significantly modified in siIRF6-depleted differentiating NHKs
siIRF6 Up-regulated Genes Gene P value HIST1H2BM histone cluster 1, H2bm 0.009024 Runx1 runt-related transcription factor 1 0.003781 zfp36l1 zinc finger protein 36, C3H type-like 1 0.010008 thbs2 thrombospondin 2 0.012696 metallothionein 1L (gene/pseudogene); metallothionein 1E; metallothionein 1 MT1L pseudogene 3; metallothionein 1J (pseudogene) 0.013351 ZFP36L2 zinc finger protein 36, C3H type-like 2 0.011236 SULF2 sulfatase 2 0.016012 FBXW5 F-box and WD repeat domain containing 5 0.017760 Bak1 BCL2-antagonist/killer 1; BCL2-like 7 pseudogene 1 0.012326 emp3 epithelial membrane protein 3 0.007449 PRKCDBP protein kinase C, delta binding protein 0.009721 rabac1 Rab acceptor 1 (prenylated) 0.025085 ITGA3 integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) 0.002824 Otud1 OTU domain containing 1 0.004225 MMP14 matrix metallopeptidase 14 (membrane-inserted) 0.028212 SNORD104 small nucleolar RNA, C/D box 104 0.034095 serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), SERPINE1 member 1 0.007105 Fat2 FAT tumor suppressor homolog 2 (Drosophila) 0.019000 VEGFC vascular endothelial growth factor C 0.021947 GRK5 G protein-coupled receptor kinase 5 0.031680 ARMC7 armadillo repeat containing 7 0.009671 Gyltl1b glycosyltransferase-like 1B 0.025608 tgfbi transforming growth factor, beta-induced, 68kDa 0.049949 TTBK2 tau tubulin kinase 2 0.017545 KIRREL kin of IRRE like (Drosophila) 0.034915 NXNL2 nucleoredoxin-like 2 0.046172 Tram2 translocation associated membrane protein 2 0.018546 DEPDC7 DEP domain containing 7 0.032088 AC106785.9 Novel rRNA pseudogene 0.042046 FAM83C family with sequence similarity 83, member C 0.041854 OVOL2 ovo-like 2 (Drosophila) 0.027120 Ngf nerve growth factor (beta polypeptide) 0.027377 Ndufb7 NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7, 18kDa 0.030164 F2R coagulation factor II (thrombin) receptor 0.013500 Lpxn leupaxin 0.026963 MAP1B microtubule-associated protein 1B 0.037878 Nnat neuronatin 0.028409 Fzd8 frizzled homolog 8 (Drosophila) 0.031618 SPARC secreted protein, acidic, cysteine-rich (osteonectin) 0.008134 Oas3 2'-5'-oligoadenylate synthetase 3, 100kDa 0.043399 MRPS26 mitochondrial ribosomal protein S26 0.006209 PTHLH parathyroid hormone-like hormone 0.001375 FLRT2 fibronectin leucine rich transmembrane protein 2 0.040131 OSBPL2 oxysterol binding protein-like 2 0.023241 HIST1H2BO histone cluster 1, H2bo 0.029435 TNC tenascin C 0.000472 NAV1 neuron navigator 1 0.032688 TMEM219 transmembrane protein 219 0.013368 AC106785.22 0.041693 LAPTM5 lysosomal multispanning membrane protein 5 0.040975 CYFIP2 cytoplasmic FMR1 interacting protein 2 0.013076 Fads3 fatty acid desaturase 3 0.026824 ARL6IP5 ADP-ribosylation-like factor 6 interacting protein 5 0.019801 Ifitm1 interferon induced transmembrane protein 1 (9-27) 0.021931 FOXD4 forkhead box D4 0.019697 IFI6 interferon, alpha-inducible protein 6 0.018705 ca12 carbonic anhydrase XII 0.009150 Sp100 SP100 nuclear antigen 0.036801 Id3 inhibitor of DNA binding 3, dominant negative helix-loop-helix protein 0.000584 NEFL neurofilament, light polypeptide 0.031607 SP110 SP110 nuclear body protein 0.009339 MXRA5 matrix-remodelling associated 5 0.001590 Cxcl14 chemokine (C-X-C motif) ligand 14 0.002662
siIRF6 Down-regulated Genes Gene P value homeodomain interacting protein kinase 2; similar to homeodomain interacting HIPK2 protein kinase 2 0.009341 KLK5 kallikrein-related peptidase 5 0.011491 TSC22D2 TSC22 domain family, member 2 0.012631 EMP2 epithelial membrane protein 2 0.011301 Hmox2 heme oxygenase (decycling) 2 0.007506 ndrg1 N-myc downstream regulated 1 0.007885 LYPD3 LY6/PLAUR domain containing 3 0.007958 DUOX1 dual oxidase 1 0.008922 POLR2A polymerase (RNA) II (DNA directed) polypeptide A, 220kDa 0.011279 LOC152845 pleiomorphic adenoma gene-like 2; similar to pleiomorphic adenoma gene-like 2 0.025148 B4galt5 UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 0.018679 KIAA1539 KIAA1539 0.024453 NPAS2 neuronal PAS domain protein 2 0.021919 Rab11a RAB11A, member RAS oncogene family 0.023472 ier2 immediate early response 2 0.006266 RHOD ras homolog gene family, member D 0.009478 Atf6 activating transcription factor 6 0.014456 cyb5r2 cytochrome b5 reductase 2 0.022777 XG Xg blood group 0.013712 Reps1 RALBP1 associated Eps domain containing 1 0.026576 ATXN2 ataxin 2 0.026946 GATA3 GATA binding protein 3 0.015595 KLF3 Kruppel-like factor 3 (basic) 0.018144 DCP1A DCP1 decapping enzyme homolog A (S. cerevisiae) 0.030803 LCE1D late cornified envelope 1D 0.019047 SIK3 serine/threonine-protein kinase QSK 0.043683 ereg epiregulin 0.004493 slc9a1 solute carrier family 9 (sodium/hydrogen exchanger), member 1 0.005644 SGPP2 sphingosine-1-phosphate phosphotase 2 0.015546 ARHGAP32 Rho GTPase-activating protein 0.022521 Dazap2 DAZ associated protein 2 0.024569 N4BP1 NEDD4 binding protein 1 0.047462 lypd6b LY6/PLAUR domain containing 6B 0.001709 IDSP1 iduronate 2-sulfatase pseudogene 1 0.018528 RAG1AP1 recombination activating gene 1 activating protein 1 0.026354 AFF1 AF4/FMR2 family, member 1 0.029775 CYB5R1 cytochrome b5 reductase 1 0.003819 crabp2 cellular retinoic acid binding protein 2 0.006792 CTTNBP2NL CTTNBP2 N-terminal like 0.009279 AVPI1 arginine vasopressin-induced 1 0.012599 STARD5 StAR-related lipid transfer (START) domain containing 5 0.015310 UBAP1 ubiquitin associated protein 1 0.023783 FAM102A family with sequence similarity 102, member A 0.010744 FAM96B family with sequence similarity 96, member B 0.003059 asap1 ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 0.003216 RAF1 v-raf-1 murine leukemia viral oncogene homolog 1 0.016222 trim8 tripartite motif-containing 8 0.028701 tinf2 TERF1 (TRF1)-interacting nuclear factor 2 0.043096 TRIB1 tribbles homolog 1 (Drosophila) 0.045878 PRSS8 protease, serine, 8 0.000930 HLA-G major histocompatibility complex, class I, G 0.013622 Cited2 Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 0.016983 KCNK1 potassium channel, subfamily K, member 1 0.021446 MTF1 metal-regulatory transcription factor 1 0.002538 ITPRIP inositol 1,4,5-triphosphate receptor interacting protein 0.018476 ACSL1 acyl-CoA synthetase long-chain family member 1 0.025941 mid1ip1 MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) 0.027438 mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase; LOC151162 hypothetical LOC151162 0.039717 siIRF6 expression profiling genes also present in ChIP-seq analysis anxa11 mpzl3 Atf6 MTF1 B4galt5 MUC15 Cited2 NAV1 Clcn5 Otud1 Clic5 OVOL1 Cxcl14 Pdzd2 Ehf PRDM1 FLRT2 PTHLH GALNT5 PXDN GATA3 RAB3IP GGT6 Raet1e GPR110 RAG1AP1 GRAMD3 Reps1 GRHL3 RYBP GRK5 SGPP2 H2AFY SLC28A3 HIPK2 SLC44A2 Hipk3 SLC5A1 irf6 Slit2 Klf4 ST6GALNAC5 KRT78 Stk40 LCE1D SULT2B1 Lmo7 TACSTD2 Lypd5 TNFAIP3 Lypd6b vgll3 MAGI1 malL MAPRE2 mid1ip1