Supporting Information
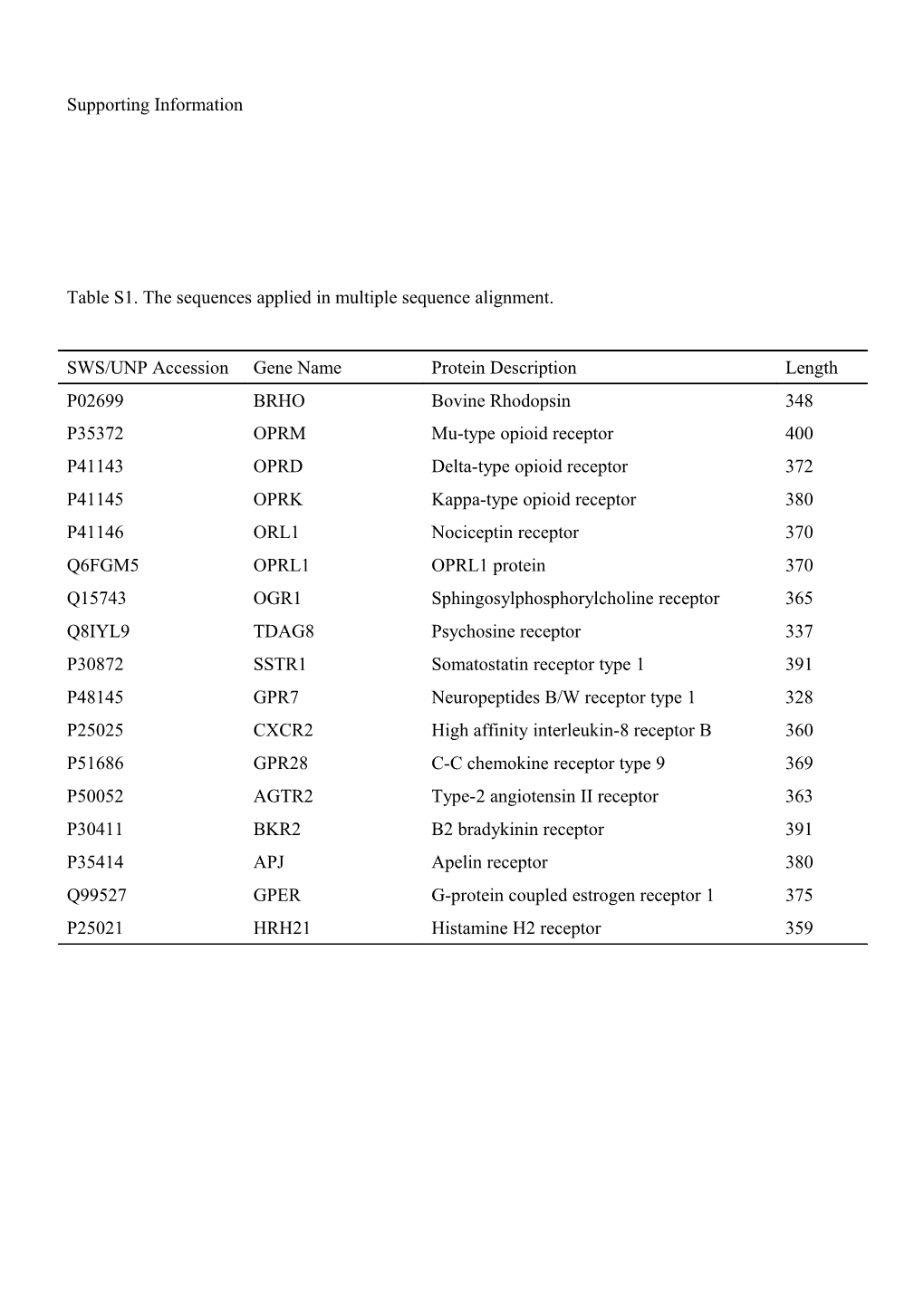
Table S1. The sequences applied in multiple sequence alignment.
SWS/UNP Accession Gene Name Protein Description Length P02699 BRHO Bovine Rhodopsin 348 P35372 OPRM Mu-type opioid receptor 400 P41143 OPRD Delta-type opioid receptor 372 P41145 OPRK Kappa-type opioid receptor 380 P41146 ORL1 Nociceptin receptor 370 Q6FGM5 OPRL1 OPRL1 protein 370 Q15743 OGR1 Sphingosylphosphorylcholine receptor 365 Q8IYL9 TDAG8 Psychosine receptor 337 P30872 SSTR1 Somatostatin receptor type 1 391 P48145 GPR7 Neuropeptides B/W receptor type 1 328 P25025 CXCR2 High affinity interleukin-8 receptor B 360 P51686 GPR28 C-C chemokine receptor type 9 369 P50052 AGTR2 Type-2 angiotensin II receptor 363 P30411 BKR2 B2 bradykinin receptor 391 P35414 APJ Apelin receptor 380 Q99527 GPER G-protein coupled estrogen receptor 1 375 P25021 HRH21 Histamine H2 receptor 359 Figure S2. Multiple sequence alignment. The transmembrane (TM) domains and Helix8 (H8) are colored in cyan, highly conserved residues are colored in red. Table S3. The template used for building up the extracellular3 (EC3), intracellular3 (IC3) and terminal regions of μ and δ receptors.
PDB ID Description Sequence Target Crystal structure of an alpha/beta hydrolase superfamily 2QRU 229-238 μ-EC3 protein from enterococcus faecalis 1J5Y Hpothetical transferase structure from thermus thermophilus 22-34 μ-IC3 137-186, 1K1G Splicing factor 1 μ-Nterm Chain A 2ENK Zinc transporter 9 30-70 μ-Cterm
2DPW Hpothetical transferase structure from thermus thermophilus 222-232 δ-EC3
1J5Y Transcriptional regulator, Biotin repressor 22-34 δ-IC3 15-44, 1CEE GTP-binding RHO-like protein δ-Nterm Chain B 2E71 Transcription elongation regulator 1 760-792 δ-Cterm Figure S4. Alignments between our receptors and the templates mentions in S3. The information about which residues match is shown below each block of residues: "*" means that the residues in that column are identical in all sequences in the alignment. ":" means that conserved substitutions have been observed. "." means that semi-conserved substitutions are observed. Figure S5. Energy changes along MD simulations for the two monomers. (a) μ receptor system, (b)δ receptor system
(a) (b)
Figure S6: Simulation time vs rmsd of the Cα of the systems with selected contact dimer. (a) μ(TM1,7)- δ(TM4,5) model1, (b) μ(TM1,7)-δ(TM4,5) model2, (c) μ(TM6,7)-δ(TM4,5) model1 and (d) μ(TM6,7)- δ(TM4,5) model2.
(a) (b)
(c) (d) S7: the coordinates of μ(TM1,7)-δ(TM4,5) contact dimer.
S8: the coordinates of μ(TM6,7)-δ(TM4,5) contact dimer.