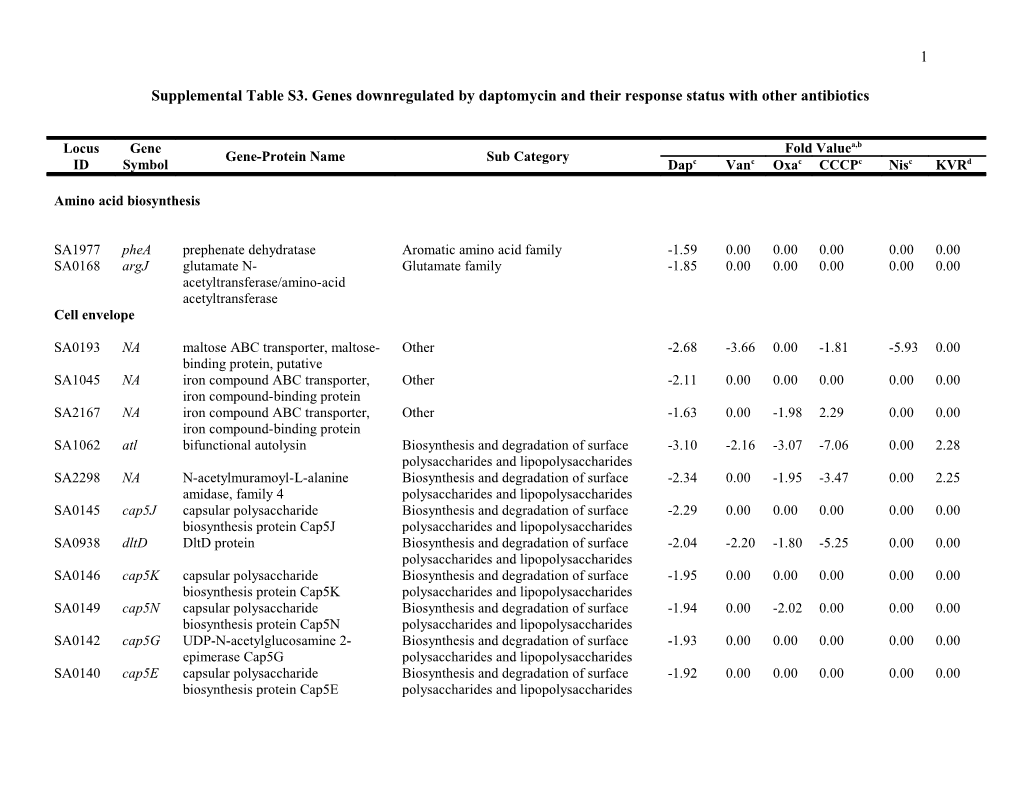
1
Supplemental Table S3. Genes downregulated by daptomycin and their response status with other antibiotics
Locus Gene Fold Valuea,b Gene-Protein Name Sub Category ID Symbol Dapc Vanc Oxac CCCPc Nisc KVRd
Amino acid biosynthesis
SA1977 pheA prephenate dehydratase Aromatic amino acid family -1.59 0.00 0.00 0.00 0.00 0.00 SA0168 argJ glutamate N- Glutamate family -1.85 0.00 0.00 0.00 0.00 0.00 acetyltransferase/amino-acid acetyltransferase Cell envelope
SA0193 NA maltose ABC transporter, maltose- Other -2.68 -3.66 0.00 -1.81 -5.93 0.00 binding protein, putative SA1045 NA iron compound ABC transporter, Other -2.11 0.00 0.00 0.00 0.00 0.00 iron compound-binding protein SA2167 NA iron compound ABC transporter, Other -1.63 0.00 -1.98 2.29 0.00 0.00 iron compound-binding protein SA1062 atl bifunctional autolysin Biosynthesis and degradation of surface -3.10 -2.16 -3.07 -7.06 0.00 2.28 polysaccharides and lipopolysaccharides SA2298 NA N-acetylmuramoyl-L-alanine Biosynthesis and degradation of surface -2.34 0.00 -1.95 -3.47 0.00 2.25 amidase, family 4 polysaccharides and lipopolysaccharides SA0145 cap5J capsular polysaccharide Biosynthesis and degradation of surface -2.29 0.00 0.00 0.00 0.00 0.00 biosynthesis protein Cap5J polysaccharides and lipopolysaccharides SA0938 dltD DltD protein Biosynthesis and degradation of surface -2.04 -2.20 -1.80 -5.25 0.00 0.00 polysaccharides and lipopolysaccharides SA0146 cap5K capsular polysaccharide Biosynthesis and degradation of surface -1.95 0.00 0.00 0.00 0.00 0.00 biosynthesis protein Cap5K polysaccharides and lipopolysaccharides SA0149 cap5N capsular polysaccharide Biosynthesis and degradation of surface -1.94 0.00 -2.02 0.00 0.00 0.00 biosynthesis protein Cap5N polysaccharides and lipopolysaccharides SA0142 cap5G UDP-N-acetylglucosamine 2- Biosynthesis and degradation of surface -1.93 0.00 0.00 0.00 0.00 0.00 epimerase Cap5G polysaccharides and lipopolysaccharides SA0140 cap5E capsular polysaccharide Biosynthesis and degradation of surface -1.92 0.00 0.00 0.00 0.00 0.00 biosynthesis protein Cap5E polysaccharides and lipopolysaccharides 2
SA0148 cap5M capsular polysaccharide Biosynthesis and degradation of surface -1.90 0.00 -1.58 0.00 0.00 0.00 biosynthesis galactosyltransferase polysaccharides and lipopolysaccharides Cap5M SA0138 cap5C capsular polysaccharide Biosynthesis and degradation of surface -1.88 0.00 -1.52 2.01 0.00 0.00 biosynthesis protein Cap5C polysaccharides and lipopolysaccharides SA0151 cap5P UDP-N-acetylglucosamine 2- Biosynthesis and degradation of surface -1.86 0.00 0.00 0.00 0.00 0.00 epimerase Cap5P polysaccharides and lipopolysaccharides SA0936 dltB DltB protein Biosynthesis and degradation of surface -1.77 0.00 -2.04 -8.60 -2.25 2.87 polysaccharides and lipopolysaccharides SA0141 cap5F capsular polysaccharide Biosynthesis and degradation of surface -1.70 0.00 0.00 -1.53 0.00 0.00 biosynthesis protein Cap5F polysaccharides and lipopolysaccharides SA0147 cap5L capsular polysaccharide Biosynthesis and degradation of surface -1.63 0.00 0.00 -1.90 0.00 0.00 biosynthesis protein Cap5L polysaccharides and lipopolysaccharides SA0137 cap5B capsular polysaccharide Biosynthesis and degradation of surface -1.61 0.00 -2.01 1.58 0.00 0.00 biosynthesis protein Cap5B polysaccharides and lipopolysaccharides SA0143 cap5H capsular polysaccharide Biosynthesis and degradation of surface -1.59 0.00 0.00 0.00 0.00 0.00 biosynthesis protein Cap5H polysaccharides and lipopolysaccharides SA0150 cap5O capsular polysaccharide Biosynthesis and degradation of surface -1.53 0.00 0.00 0.00 0.00 0.00 biosynthesis protein Cap5O polysaccharides and lipopolysaccharides SA0139 cap5D capsular polysaccharide Biosynthesis and degradation of surface -1.53 0.00 -1.85 1.65 0.00 0.00 biosynthesis protein Cap5D polysaccharides and lipopolysaccharides SA1329 femC glutamine synthetase FemC Biosynthesis of murein sacculus and -2.14 0.00 0.00 -2.44 0.00 0.00 peptidoglycan SA0245 lytS sensor histidine kinase LytS Biosynthesis of murein sacculus and -1.70 0.00 -1.69 2.21 2.05 0.00 peptidoglycan SA1609 pbp3 penicillin-binding protein 3 Biosynthesis of murein sacculus and -1.66 0.00 0.00 -1.75 -1.90 0.00 peptidoglycan SA1195 mraY phospho-N-acetylmuramoyl- Biosynthesis of murein sacculus and -1.56 -1.80 0.00 -4.88 0.00 0.00 pentapeptide-transferase peptidoglycan SA0193 NA maltose ABC transporter, maltose- Other -2.68 -3.66 0.00 -1.81 -5.93 0.00 binding protein, putative SA2409 fmhA fmhA protein Other -2.56 0.00 0.00 -2.37 0.00 0.00 SA1806 sasC probable ATP-dependent helicase Other -2.29 -1.77 0.00 0.00 0.00 0.00 SA2582 NA membrane protein, putative Other -2.15 0.00 0.00 -4.88 -1.95 1.63 SA1045 NA iron compound ABC transporter, Other -2.11 0.00 0.00 0.00 0.00 0.00 iron compound-binding protein SA2676 sasA LPXTG cell wall surface anchor Other -1.87 0.00 1.75 -3.29 0.00 1.85 3
family protein SA1140 sasE LPXTG cell wall surface anchor Other -1.77 0.00 0.00 0.00 0.00 0.00 protein SA0411 NA membrane protein, putative Other -1.72 0.00 0.00 -2.68 0.00 0.00 SA0189 NA hypothetical protein Other -1.68 0.00 0.00 0.00 0.00 0.00 SA2167 NA iron compound ABC transporter, Other -1.63 0.00 -1.98 2.29 0.00 0.00 iron compound-binding protein SA2407 NA lipoprotein, putative Other -1.59 0.00 0.00 0.00 0.00 0.00 SA2505 sasG cell wall surface anchor family Other -1.56 0.00 -1.97 2.08 0.00 0.00 protein SA2082 NA membrane protein, putative Other -1.54 0.00 0.00 -8.74 0.00 2.80 SA2272 modA molybdenum ABC transporter, Other -1.53 0.00 -2.08 2.39 0.00 0.00 molybdenum-binding protein ModA SA2409 fmhA fmhA protein Other -2.56 0.00 0.00 -2.37 0.00 0.00
Cellular processes
SA2437 bcr bicyclomycin resistance protein Toxin production and resistance -1.86 0.00 0.00 -3.75 -1.84 0.00 SA2460 NA drug transporter, putative Toxin production and resistance -1.74 0.00 0.00 -2.97 0.00 0.00 SA2257 NA drug transporter, putative Toxin production and resistance -1.73 0.00 0.00 -3.10 0.00 0.00 SA2347 NA drug resistance transporter, Toxin production and resistance -1.57 0.00 -1.66 -1.72 0.00 1.56 EmrB/QacA subfamily SA2054 rpoF RNA polymerase sigma-37 factor Adaptations to atypical conditions -1.77 0.00 0.00 -6.13 0.00 0.00 SA1970 sspB2 cysteine protease precursor SspB Pathogenesis -1.81 0.00 0.00 -2.09 0.00 0.00 SA1328 glnR glutamine synthetase repressor Toxin production and resistance -2.74 -2.59 -2.30 -2.29 0.00 0.00 SA2025 argC2 accessory gene regulator protein C Pathogenesis -1.91 0.00 0.00 -1.90 -1.95 0.00 SA2409 fmhA fmhA protein Toxin production and resistance -2.56 -2.40 -2.60 -2.37 0.00 1.90 SA2627 betA choline dehydrogenase Adaptations to atypical conditions -2.51 -2.20 -1.80 -2.68 0.00 2.30 SA1437 NA cold shock protein, CSD family Adaptations to atypical conditions -2.17 -2.90 -2.95 -2.65 0.00 2.50 SA1753 NA universal stress protein family Adaptations to atypical conditions -1.99 0.00 -1.52 2.00 0.00 0.00 SA0861 NA cold shock protein, CSD family Adaptations to atypical conditions -1.95 0.00 0.00 -1.87 0.00 0.00 SA2731 NA cold shock protein, CSD family Adaptations to atypical conditions -1.77 0.00 0.00 -2.32 0.00 0.00 SA2054 rpoF RNA polymerase sigma-37 factor Adaptations to atypical conditions -1.77 0.00 0.00 -6.13 0.00 0.00 SA0609 sdrD sdrD protein Cell adhesion -2.71 0.00 -2.32 0.00 0.00 2.60 SA2738 trmE tRNA modification GTPase TrmE Detoxification -1.61 0.00 0.00 0.00 0.00 0.00 SA0270 NA staphyloxanthin biosynthesis Pathogenesis -7.13 -1.85 0.00 -6.30 0.00 0.00 4
protein, putative SA2291 NA staphyloxanthin biosynthesis Pathogenesis -4.15 -3.08 -2.07 -13.34 -3.74 6.54 protein SA2581 NA staphyloxanthin biosynthesis Pathogenesis -3.55 0.00 0.00 -3.17 0.00 0.00 protein SA0095 spa immunoglobulin G binding protein Pathogenesis -2.77 0.00 0.00 1.93 0.00 0.00 A precursor SA2652 clfB clumping factor B Pathogenesis -2.50 0.00 0.00 -5.94 0.00 1.67 SA2023 agrB accessory gene regulator protein B Pathogenesis -1.96 -1.87 -1.61 -2.21 0.00 0.00 SA2025 argC2 accessory gene regulator protein C Pathogenesis -1.91 -1.90 -1.78 -1.90 -1.95 0.00 SA1970 sspB2 cysteine protease precursor SspB Pathogenesis -1.81 0.00 0.00 -2.09 0.00 0.00 SA1184 NA exfoliative toxin, putative Pathogenesis -1.78 0.00 0.00 -1.61 0.00 0.00 SA2287 sarR staphylococcal accessory regulator Pathogenesis -1.64 -1.80 -1.73 0.00 0.00 0.00 R SA0096 sarS staphylococcal accessory regulator Toxin production and resistance -2.81 -1.90 -1.86 0.00 0.00 0.00 S SA1328 glnR glutamine synthetase repressor Toxin production and resistance -2.74 -2.59 0.00 -2.29 0.00 0.00 SA2409 fmhA fmhA protein Toxin production and resistance -2.56 0.00 0.00 -2.37 0.00 0.00 SA0935 dltA D-alanine-activating enzyme/D- Toxin production and resistance -2.13 0.00 -2.09 -4.78 -2.16 1.75 alanine-D-alanyl carrier protein ligase SA2437 bcr bicyclomycin resistance protein Toxin production and resistance -1.86 0.00 0.00 -3.75 -1.84 0.00 SA0746 norR transcriptional regulator, MarR Toxin production and resistance -1.80 1.91 -1.53 -6.32 -1.58 0.00 family SA1184 NA exfoliative toxin, putative Toxin production and resistance -1.78 0.00 0.00 -1.61 0.00 0.00 SA2460 NA drug transporter, putative Toxin production and resistance -1.74 0.00 0.00 -2.97 0.00 0.00 SA2257 NA drug transporter, putative Toxin production and resistance -1.73 0.00 0.00 -3.10 0.00 0.00 SA1173 hlY alpha-hemolysin precursor Toxin production and resistance -1.66 0.00 0.00 0.00 0.00 0.00 SA1152 NA colicin V production protein, Toxin production and resistance -1.57 0.00 0.00 0.00 0.00 0.00 putative SA2347 NA drug resistance transporter, Toxin production and resistance -1.57 0.00 -1.66 -1.72 0.00 1.56 EmrB/QacA subfamily SA1812 rot repressor of toxins Toxin production and resistance -1.56 -1.78 -1.54 0.00 0.00 1.70 SA1396 fmtC fmtC protein Toxin production and resistance -1.54 -2.80 -2.19 0.00 -1.94 0.00
Central intermediary metabolism 5
SA2395 narG respiratory nitrate reductase, alpha Nitrogen metabolism -1.55 0.00 0.00 0.00 11.95 0.00 subunit SA1837 metK S-adenosylmethionine synthetase One-carbon metabolism -1.55 0.00 0.00 -5.43 0.00 1.83 SA1760 ackA acetate kinase Other -2.05 0.00 -1.84 -2.10 1.97 1.56 SA2535 NA D-isomer specific 2-hydroxyacid Other -1.75 0.00 0.00 -5.12 4.59 0.00 dehydrogenase family protein SA1976 NA nitric-oxide synthase, oxygenase Other -1.61 0.00 0.00 0.00 0.00 1.52 subunit SA1760 ackA acetate kinase Other -2.05 0.00 -1.84 -2.10 1.97 1.56
DNA metabolism
SA1357 NA thermonuclease precursor family Degradation of DNA -1.61 0.00 0.00 0.00 0.00 0.00 protein SA0438 ssb2 single-stranded DNA-binding DNA replication, recombination, and repair -2.53 0.00 0.00 -1.89 0.00 2.30 protein SA1691 recJ single-stranded-DNA-specific DNA replication, recombination, and repair -2.12 0.00 0.00 0.00 0.00 0.00 exonuclease RecJ SA1614 nfo endonuclease IV DNA replication, recombination, and repair -1.95 0.00 0.00 0.00 0.00 0.00 SA0627 ung uracil-DNA glycosylase DNA replication, recombination, and repair -1.77 0.00 0.00 0.00 0.00 0.00 SA1374 lexA LexA repressor DNA replication, recombination, and repair -1.70 0.00 0.00 0.00 0.00 0.00 SA0526 NA DNA polymerase III, delta prime DNA replication, recombination, and repair -1.61 0.00 0.00 0.00 0.00 0.00 subunit, putative SA0180 NA type I restriction-modification Restriction/modification -2.35 -1.64 0.00 -2.58 -1.83 0.00 enzyme, R subunit SA0477 NA type I restriction-modification Restriction/modification -1.74 0.00 0.00 0.00 0.00 0.00 system, S subunit, EcoA family, putative SA0476 hsdM1 type I restriction-modification Restriction/modification -1.54 0.00 -1.51 0.00 -1.99 0.00 system, M subunit
Energy metabolism
SA2395 narG respiratory nitrate reductase, alpha Anaerobic -1.55 0.00 0.00 0.00 11.95 0.00 subunit SA1760 ackA acetate kinase Fermentation -2.05 -2.30 -1.84 -2.10 1.97 1.56 6
SA0008 hutH histidine ammonia-lyase Amino acids and amines -1.73 0.00 0.00 -1.67 -3.55 0.00 SA2545 sdaAB L-serine dehydratase, iron-sulfur- Amino acids and amines -1.56 -1.60 0.00 -3.08 0.00 0.00 dependent, beta subunit SA2544 sdaAA L-serine dehydratase, iron-sulfur- Amino acids and amines -1.51 -1.63 0.00 -3.15 -2.37 0.00 dependent, alpha subunit SA2301 NA formate dehydrogenase, alpha Anaerobic -1.66 0.00 -2.72 0.00 0.00 -2.14 subunit, putative SA2395 narG respiratory nitrate reductase, alpha Anaerobic -1.55 0.00 0.00 0.00 11.95 0.00 subunit SA0517 treC alpha-amylase family protein Biosynthesis and degradation of -1.53 0.00 0.00 0.00 0.00 0.00 polysaccharides SA1068 qoxC quinol oxidase, subunit III Electron transport -4.18 -2.68 -2.27 0.00 0.00 0.00 SA1067 qoxD quinol oxidase, subunit IV Electron transport -3.10 -2.50 -2.67 0.00 0.00 0.00 SA1069 qoxA quinol oxidase, subunit I Electron transport -2.31 -2.04 -3.04 0.00 0.00 0.00 SA1070 qoxB quinol oxidase, subunit II Electron transport -2.24 -1.52 -2.85 0.00 0.00 2.47 SA0190 NA NAD(P)H dehydrogenase Electron transport -1.65 0.00 0.00 2.30 0.00 0.00 (quinone), putative SA1914 NA iron-sulfur cluster-binding protein, Electron transport -1.63 0.00 0.00 -1.73 -3.01 2.68 putative SA1525 fer ferredoxin Electron transport -1.52 0.00 0.00 0.00 0.00 0.00 SA1760 ackA acetate kinase Fermentation -2.05 0.00 -1.84 -2.10 1.97 1.56 SA1734 gapA2 glyceraldehyde 3-phosphate Glycolysis/gluconeogenesis -3.27 -1.92 0.00 -2.23 -5.63 2.62 dehydrogenase SA1320 glpK glycerol kinase Other -1.64 -1.56 0.00 -5.83 -3.98 1.86 SA2329 rpiA ribose 5-phosphate isomerase Pentose phosphate pathway -1.81 0.00 0.00 0.00 0.00 0.00 SA2135 manA1 mannose-6-phosphate isomerase, Sugars -2.47 -2.40 -1.59 -6.41 -4.29 2.67 class I SA2664 manA2 mannose-6-phosphate isomerase Sugars -1.67 -2.24 -1.76 0.00 -1.79 0.00 SA2332 galM aldose 1-epimerase Sugars -1.62 0.00 0.00 2.19 0.00 0.00 SA1160 sdhB succinate dehydrogenase, iron- TCA cycle -1.81 -1.72 0.00 -2.75 -1.75 0.00 sulfur protein SA1158 sdhC succinate dehydrogenase, TCA cycle -1.65 -1.73 0.00 -2.98 -1.78 0.00 cytochrome b558 subunit SA2623 mqo2 malate:quinone oxidoreductase TCA cycle -1.51 0.00 -1.62 0.00 0.00 0.00
Fatty acid and phospholipid metabolism 7
SA1661 NA acetyl-CoA carboxylase, biotin Biosynthesis -2.07 -1.93 0.00 0.00 -3.56 0.00 carboxylase, putative SA1662 NA acetyl-CoA carboxylase, biotin Biosynthesis -1.94 0.00 0.00 0.00 0.00 0.00 carboxyl carrier protein, putative SA1747 accA acetyl-CoA carboxylase, carboxyl Biosynthesis -1.57 0.00 0.00 0.00 0.00 0.00 transferase, alpha subunit SA0317 NA lipase precursor, interruption Degradation -3.27 -2.53 0.00 0.00 0.00 0.00 SA0078 plc 1-phosphatidylinositol Degradation -1.60 0.00 0.00 0.00 0.00 0.00 phosphodiesterase
Protein fate
SA1667 NA peptidase, U32 family Degradation of proteins, peptides, and -1.93 -1.80 -1.79 -3.70 -1.77 1.77 glycopeptides SA1036 NA protease, putative Degradation of proteins, peptides, and -1.83 0.00 0.00 -1.81 0.00 0.00 glycopeptides SA1970 sspB2 cysteine protease precursor SspB Degradation of proteins, peptides, and -1.81 0.00 0.00 -2.09 0.00 0.00 glycopeptides SA1756 pepQ truncated amidase Degradation of proteins, peptides, and -1.77 0.00 0.00 -1.72 0.00 0.00 glycopeptides SA1692 NA protein-export membrane protein Protein and peptide secretion and -1.68 0.00 0.00 -4.52 -2.11 1.67 SecDF trafficking SA2675 NA SecY/Sec61-alpha family protein Protein and peptide secretion and -1.67 0.00 0.00 0.00 0.00 0.00 trafficking SA1208 lspA lipoprotein signal peptidase Protein and peptide secretion and -1.59 0.00 0.00 -1.65 0.00 0.00 trafficking SA1251 NA cell division protein FtsY, putative Protein and peptide secretion and -1.51 0.00 0.00 -1.59 0.00 0.00 trafficking SA1722 tig trigger factor Protein folding and stabilization -1.57 1.53 0.00 0.00 0.00 1.92 SA1227 def2 polypeptide deformylase Protein modification and repair -1.71 0.00 0.00 0.00 0.00 0.00 SA1970 sspB2 cysteine protease precursor SspB Degradation of proteins, peptides, and -1.81 0.00 0.00 -2.09 0.00 0.00 glycopeptides
Protein synthesis
SA1726 rpmI ribosomal protein L35 Ribosomal proteins: synthesis and -3.05 -2.80 -2.50 -3.83 0.00 0.00 8
modification SA0585 rplJ ribosomal protein L10 Ribosomal proteins: synthesis and -2.65 -1.76 -1.84 -5.62 -2.27 2.50 modification SA1725 rplT ribosomal protein L20 Ribosomal proteins: synthesis and -2.61 -2.16 0.00 -6.54 0.00 3.73 modification SA0586 rplL ribosomal protein L7/L12 Ribosomal proteins: synthesis and -2.55 -2.12 -1.71 -7.84 -1.63 1.72 modification SA1254 rpsP ribosomal protein S16 Ribosomal proteins: synthesis and -2.55 0.00 0.00 -3.79 0.00 1.65 modification SA1769 rpsD ribosomal protein S4 Ribosomal proteins: synthesis and -2.43 -1.96 0.00 -3.20 -2.02 2.76 modification SA0439 rpsR ribosomal protein S18 Ribosomal proteins: synthesis and -2.31 0.00 0.00 -4.40 0.00 0.00 modification SA2224 rplF ribosomal protein L6 Ribosomal proteins: synthesis and -2.30 0.00 0.00 -2.28 0.00 1.86 modification SA0590 NA 30S ribosomal protein L7 Ae Ribosomal proteins: synthesis and -2.18 0.00 0.00 -6.05 0.00 0.00 modification SA2233 rpsC ribosomal protein S3 Ribosomal proteins: synthesis and -2.05 -1.95 0.00 -3.71 -3.58 2.53 modification SA2740 rpmH ribosomal protein L34 Ribosomal proteins: synthesis and -2.02 0.00 0.00 -2.50 0.00 0.00 modification SA0437 rpsF ribosomal protein S6 Ribosomal proteins: synthesis and -2.02 -1.63 0.00 -5.08 -1.50 3.13 modification SA2215 rpsM ribosomal protein S13/S18 Ribosomal proteins: synthesis and -2.01 -1.55 0.00 -2.54 -1.50 1.61 modification SA1642 rpsT ribosomal protein S20 Ribosomal proteins: synthesis and -1.98 0.00 0.00 -3.44 0.00 0.00 modification SA2236 rplB ribosomal protein L2 Ribosomal proteins: synthesis and -1.95 -1.71 0.00 -4.05 0.00 2.87 modification SA1137 rpmF ribosomal protein L32 Ribosomal proteins: synthesis and -1.93 -1.66 0.00 0.00 0.00 0.00 modification SA2232 rplP ribosomal protein L16 Ribosomal proteins: synthesis and -1.93 -1.72 0.00 -2.45 0.00 0.00 modification SA2228 rplX ribosomal protein L24 Ribosomal proteins: synthesis and -1.90 0.00 0.00 -3.08 0.00 0.00 modification SA1257 rplS ribosomal protein L19 Ribosomal proteins: synthesis and -1.89 0.00 0.00 -4.21 -1.76 0.00 modification 9
SA0592 rpsG ribosomal protein S7 Ribosomal proteins: synthesis and -1.81 -1.84 -1.75 -7.36 -1.70 1.52 modification SA2223 rplR ribosomal protein L18 Ribosomal proteins: synthesis and -1.81 -1.52 0.00 -2.59 0.00 1.86 modification SA2225 rpsH ribosomal protein S8 Ribosomal proteins: synthesis and -1.80 -1.56 0.00 -2.62 0.00 1.66 modification SA1700 rpmA ribosomal protein L27 Ribosomal proteins: synthesis and -1.78 0.00 0.00 0.00 0.00 1.59 modification SA2230 rpsQ ribosomal protein S17 Ribosomal proteins: synthesis and -1.78 0.00 0.00 -2.11 0.00 0.00 modification SA1238 rpmB ribosomal protein L28 Ribosomal proteins: synthesis and -1.75 0.00 0.00 0.00 0.00 0.00 modification SA2212 rplQ ribosomal protein L17 Ribosomal proteins: synthesis and -1.75 0.00 0.00 -2.21 0.00 0.00 modification SA1292 rpsO ribosomal protein S15 Ribosomal proteins: synthesis and -1.72 0.00 0.00 0.00 0.00 0.00 modification SA2220 rplO ribosomal protein L15 Ribosomal proteins: synthesis and -1.72 -1.51 0.00 -2.09 0.00 0.00 modification SA2235 rpsS ribosomal protein S19 Ribosomal proteins: synthesis and -1.70 -2.66 0.00 -4.45 -1.65 2.41 modification SA2237 rplW ribosomal protein L23 Ribosomal proteins: synthesis and -1.70 0.00 0.00 -5.26 -1.51 3.77 modification SA2221 rpmD ribosomal protein L30p/L7e Ribosomal proteins: synthesis and -1.69 0.00 0.00 0.00 0.00 0.00 modification SA2227 rplE ribosomal protein L5 Ribosomal proteins: synthesis and -1.67 -1.54 0.00 -2.87 0.00 2.21 modification SA1608 rpmG2 ribosomal protein L33 Ribosomal proteins: synthesis and -1.64 0.00 0.00 0.00 0.00 0.00 modification SA2240 rpsJ ribosomal protein S10 Ribosomal proteins: synthesis and -1.63 -1.67 0.00 -7.91 -2.26 3.61 modification SA2226 rpsN2 ribosomal protein S14 Ribosomal proteins: synthesis and -1.61 0.00 0.00 -1.85 0.00 1.64 modification SA0584 rplA ribosomal protein L1 Ribosomal proteins: synthesis and -1.59 0.00 0.00 -7.34 -1.50 1.56 modification SA1632 rpsU ribosomal protein S21 Ribosomal proteins: synthesis and -1.57 0.00 0.00 0.00 0.00 0.00 modification SA2112 rpmE ribosomal protein L31 Ribosomal proteins: synthesis and -1.55 0.00 0.00 0.00 0.00 0.00 10
modification SA2231 rpmC ribosomal protein L29 Ribosomal proteins: synthesis and -1.52 -1.84 0.00 -2.69 0.00 1.56 modification SA2238 rplD ribosomal protein L4 Ribosomal proteins: synthesis and -1.52 -1.71 0.00 -7.59 0.00 3.56 modification SA1727 infC translation initiation factor IF-3 Translation factors -3.01 -1.81 0.00 -8.22 -1.83 3.95 SA2110 prfA peptide chain release factor 1 Translation factors -2.27 -2.21 -1.45 -6.77 -1.59 2.01 SA1587 efp translation elongation factor P Translation factors -1.92 0.00 -1.62 -1.72 0.00 0.00 SA2217 infA translation initiation factor IF-1 Translation factors -1.80 0.00 0.00 0.00 0.00 0.00 SA0593 fusA translation elongation factor G Translation factors -1.67 0.00 0.00 -2.71 0.00 0.00 SA1025 prfC peptide chain release factor 3 Translation factors -1.60 0.00 0.00 0.00 0.00 0.00 SA1148 pheS phenylalanyl-tRNA synthetase, tRNA aminoacylation -2.26 0.00 0.00 -4.17 0.00 1.64 alpha subunit SA1729 thrS threonyl-tRNA synthetase tRNA aminoacylation -1.95 0.00 -1.87 2.42 0.00 0.00 SA1149 pheT phenylalanyl-tRNA synthetase, beta tRNA aminoacylation -1.89 0.00 0.00 -1.97 0.00 0.00 subunit SA0009 serS seryl-tRNA synthetase tRNA aminoacylation -1.87 0.00 -1.53 0.00 0.00 0.00 SA1228 fmt methionyl-tRNA formyltransferase tRNA aminoacylation -1.76 -1.74 0.00 -1.80 0.00 0.00 SA1001 trpS tryptophanyl-tRNA synthetase tRNA aminoacylation -1.65 0.00 -1.75 0.00 -1.94 0.00 SA1962 gatC glutamyl-tRNA(Gln) tRNA aminoacylation -1.58 0.00 0.00 -4.28 -4.19 1.57 amidotransferase, C subunit SA1778 tyrS tyrosyl-tRNA synthetase tRNA aminoacylation -1.57 0.00 0.00 -4.76 0.00 0.00 SA1256 trmD tRNA (guanine-N1)- tRNA and rRNA base modification -3.31 0.00 -1.60 -2.90 0.00 0.00 methyltransferase SA1907 NA ribosomal large subunit tRNA and rRNA base modification -1.84 0.00 0.00 0.00 0.00 1.73 pseudouridine synthase, RluD subfamily SA1957 NA RNA methyltransferase, TrmA tRNA and rRNA base modification -1.84 0.00 0.00 -2.64 0.00 0.00 family SA1803 NA pseudouridine synthase, family 1 tRNA and rRNA base modification -1.73 0.00 0.00 0.00 0.00 0.00
Regulatory functions
SA1107 NA transcriptional regulator, Cro/CI DNA interactions -3.34 0.00 0.00 -4.86 1.50 0.00 family SA1328 glnR glutamine synthetase repressor DNA interactions -2.74 -2.59 0.00 -2.29 0.00 0.00 SA1552 malR maltose operon repressor DNA interactions -2.10 -2.61 0.00 -3.78 -1.77 0.00 11
SA2317 NA phosphosugar-binding DNA interactions -1.94 0.00 0.00 -1.86 0.00 0.00 transcriptional regulator SA0249 NA transcriptional regulator, GntR DNA interactions -1.89 0.00 0.00 -2.14 0.00 0.00 family SA1997 NA transcriptional regulator, GntR DNA interactions -1.88 0.00 0.00 -4.43 0.00 0.00 family SA2325 NA transcriptional regulator, LysR DNA interactions -1.68 0.00 0.00 0.00 0.00 0.00 family SA1904 NA transcriptional regulator, putative DNA interactions -1.62 0.00 0.00 0.00 0.00 0.00 SA2546 NA perfringolysin O regulator protein, Other -1.91 -2.46 0.00 -4.88 -2.73 2.03 putative SA2025 argC2 accessory gene regulator protein C Other -1.91 0.00 0.00 -1.90 -1.95 0.00 SA1232 NA protein kinase. POINT Protein interactions -1.95 0.00 0.00 -4.17 -1.91 1.74 MUTATION SA2107 NA phosphotyrosine protein Protein interactions -1.64 0.00 0.00 -2.13 0.00 0.00 phosphatase SA1210 pyrR pyrimidine operon regulatory RNA interactions -3.80 -1.62 0.00 -7.54 -4.90 4.84 protein SA2025 argC2 accessory gene regulator protein C Other -1.91 0.00 0.00 -1.90 -1.95 0.00 SA1328 glnR glutamine synthetase repressor DNA interactions -2.74 -2.59 0.00 -2.29 0.00 0.00
Signal transduction
SA2316 NA PTS system, IIBC components PTS -2.43 -2.64 0.00 -8.25 -3.83 0.00 SA0175 NA PTS system, IIABC components PTS -2.27 0.00 -1.93 -3.21 0.00 0.00 SA2552 NA PTS system, IIABC components PTS -2.14 0.00 0.00 -2.13 0.00 0.00 SA2376 NA PTS system, sucrose-specific IIBC PTS -1.88 -1.81 -1.78 -10.57 -2.24 -1.61 components, putative SA0516 NA PTS system, IIBC components PTS -1.55 0.00 0.00 -3.25 0.00 -1.57 SA2181 lacE PTS system, lactose-specific IIBC PTS -1.52 0.00 0.00 1.64 0.00 0.00 components SA2663 NA PTS system, fructose-specific PTS -1.51 0.00 0.00 0.00 -3.85 0.00 IIABC components SA2316 NA PTS system, IIBC components PTS -2.43 -2.64 0.00 -8.25 -3.83 0.00 SA0175 NA PTS system, IIABC components PTS -2.27 0.00 -1.93 -3.21 0.00 0.00 SA2552 NA PTS system, IIABC components PTS -2.14 0.00 0.00 -2.13 0.00 0.00 SA2376 NA PTS system, sucrose-specific IIBC PTS -1.88 -1.81 0.00 -10.57 -2.24 -1.61 12
components, putative SA0516 NA PTS system, IIBC components PTS -1.55 0.00 0.00 -3.25 0.00 -1.57 SA2181 lacE PTS system, lactose-specific IIBC PTS -1.52 0.00 0.00 1.64 0.00 0.00 components SA2663 NA PTS system, fructose-specific PTS -1.51 0.00 0.00 0.00 -3.85 0.00 IIABC components
Transcription
SA1222 rpoZ DNA-directed RNA polymerase, DNA-dependent RNA polymerase -1.93 -1.80 -1.90 0.00 0.00 0.00 omega subunit SA2213 rpoA DNA-directed RNA polymerase, DNA-dependent RNA polymerase -1.81 -1.90 -1.70 0.00 0.00 1.71 alpha subunit SA2072 NA ATP-dependent RNA helicase, Other -2.80 0.00 0.00 -5.30 0.00 2.83 DEAD/DEAH box family SA1615 NA ATP-dependent RNA helicase, Other -1.75 0.00 0.00 -2.12 0.00 0.00 DEAD/DEAH box family SA1255 rimM 16S rRNA processing protein RNA processing -2.14 0.00 0.00 -1.83 0.00 0.00 RimM SA2739 rnpA ribonuclease P protein component RNA processing -1.67 0.00 0.00 -1.99 0.00 0.00 SA2054 rpoF RNA polymerase sigma-37 factor Transcription factors -1.77 0.00 0.00 -6.13 0.00 0.00 SA2055 rsbW anti-sigma B factor Transcription factors -1.50 0.00 -1.87 -2.66 0.00 0.00
Transport and binding proteins
SA0303 NA acid phosphatase5-nucleotidase, Other -2.87 0.00 0.00 0.00 0.00 0.00 lipoprotein e(P4) family SA0689 NA ABC transporter, permease protein Unknown substrate -5.55 0.00 0.00 -5.72 -3.23 0.00 SA0690 NA ABC transporter, ATP-binding Unknown substrate -5.25 0.00 0.00 -3.63 -8.28 2.10 protein SA0688 NA ABC transporter, substrate-binding Unknown substrate -5.20 0.00 0.00 -5.09 -3.91 0.00 protein SA2401 NA formate/nitrite transporter family Anions -3.88 0.00 -1.89 -3.06 0.00 0.00 protein SA0566 nupC nucleoside permease NupC Nucleosides, purines and pyrimidines -3.84 -2.89 0.00 -4.88 0.00 1.62 SA1108 NA spermidine/putrescine ABC Amino acids, peptides and amines -3.70 0.00 0.00 -4.75 0.00 0.00 transporter, ATP-binding protein 13
SA0192 NA maltose ABC transporter, ATP- Carbohydrates, organic alcohols, and acids -3.63 -3.52 0.00 -2.01 -9.14 2.68 binding protein, putative SA0093 NA L-lactate permease Carbohydrates, organic alcohols, and acids -3.41 -2.80 -2.78 -6.89 -1.81 1.99 SA2632 cudT osmoprotectant transporter, BCCT Amino acids, peptides and amines -3.35 0.00 0.00 -5.02 0.00 0.00 family SA1084 NA cobalt transport family protein Cations and iron carrying compounds -3.13 0.00 0.00 -3.92 0.00 0.00 SA2521 NA transporter, putative Unknown substrate -3.04 -2.30 -1.50 -6.69 -4.57 0.00 SA0701 NA nucleoside permease NupC, Nucleosides, purines and pyrimidines -2.99 -1.61 -2.02 -4.65 0.00 1.62 putative SA1110 NA spermidine/putrescine ABC Amino acids, peptides and amines -2.90 0.00 0.00 -3.22 0.00 0.00 transporter, permease protein SA0303 NA acid phosphatase5-nucleotidase, Other -2.87 0.00 0.00 0.00 0.00 0.00 lipoprotein e(P4) family SA2525 NA ABC transporter, ATP-binding Unknown substrate -2.85 0.00 -1.67 0.00 0.00 0.00 protein SA0171 brnQ1 branched-chain amino acid Amino acids, peptides and amines -2.85 0.00 -1.89 -2.92 0.00 0.00 transport system II carrier protein SA0407 glpT glycerol-3-phosphate transporter Other -2.84 0.00 0.00 -4.08 -2.92 1.93 SA0264 NA ABC transporter, ATP-binding Unknown substrate -2.71 0.00 -1.69 0.00 1.95 0.00 protein SA0193 NA maltose ABC transporter, maltose- Carbohydrates, organic alcohols, and acids -2.68 -3.66 0.00 -1.81 -5.93 0.00 binding protein, putative SA1367 NA amino acid permease Amino acids, peptides and amines -2.63 0.00 -2.36 -5.27 0.00 0.00 SA2242 NA xanthine/uracil permease family Nucleosides, purines and pyrimidines -2.56 -2.20 0.00 -15.03 -2.15 3.07 protein SA2382 NA proton/sodium-glutamate symport Amino acids, peptides and amines -2.51 0.00 0.00 -7.71 -3.41 1.68 protein SA0099 sirA iron compound ABC transporter, Cations and iron carrying compounds -2.50 0.00 0.00 -1.81 0.00 0.00 iron compound-binding protein SirA SA1085 NA ABC transporter, ATP-binding Unknown substrate -2.49 0.00 0.00 -4.21 -1.72 1.96 protein SA1111 NA spermidine/putrescine ABC Amino acids, peptides and amines -2.45 0.00 0.00 -4.44 0.00 -2.62 transporter, spermidine/putrescine- binding protein SA2316 NA PTS system, IIBC components Carbohydrates, organic alcohols, and acids -2.43 -2.64 0.00 -8.25 -3.83 0.00 SA0194 NA maltose ABC transporter, permease Carbohydrates, organic alcohols, and acids -2.41 -2.72 0.00 0.00 -5.42 0.00 14
protein SA2340 gltS sodium:glutamate symporter Amino acids, peptides and amines -2.40 0.00 0.00 -1.81 0.00 0.00 SA1952 NA ferritins family protein Cations and iron carrying compounds -2.31 0.00 0.00 0.00 3.01 0.00 SA2335 NA ABC transporter, ATP-binding Unknown substrate -2.29 0.00 -1.70 -2.78 0.00 0.00 protein SA0175 NA PTS system, IIABC components Carbohydrates, organic alcohols, and acids -2.27 0.00 -1.93 -3.21 0.00 0.00 SA0459 pbuX xanthine permease Nucleosides, purines and pyrimidines -2.24 0.00 -1.96 -6.06 -1.94 3.43 SA0620 proP osmoprotectant proline transporter Amino acids, peptides and amines -2.16 0.00 0.00 -4.13 0.00 0.00 SA2552 NA PTS system, IIABC components Carbohydrates, organic alcohols, and acids -2.14 0.00 0.00 -2.13 0.00 0.00 SA1996 NA ABC transporter, ATP-binding Unknown substrate -2.13 0.00 0.00 -6.87 -2.59 0.00 protein SA1045 NA iron compound ABC transporter, Cations and iron carrying compounds -2.11 0.00 0.00 0.00 0.00 0.00 iron compound-binding protein SA1319 glpF glycerol uptake facilitator protein Other -2.05 0.00 0.00 -1.61 0.00 2.05 SA0088 NA Na/Pi cotransporter family protein Anions -1.95 0.00 -1.93 -1.55 0.00 0.00 SA1979 NA sodium-dependent transporter Unknown substrate -1.93 0.00 0.00 -3.40 -3.01 0.00 SA0311 NA sodium:solute symporter family Cations and iron carrying compounds -1.90 0.00 0.00 0.00 0.00 0.00 protein SA0722 NA phosphate transporter family Anions -1.90 0.00 0.00 -7.39 -2.00 2.35 protein SA2376 NA PTS system, sucrose-specific IIBC Carbohydrates, organic alcohols, and acids -1.88 -1.81 0.00 -10.57 -2.24 -1.61 components, putative SA2363 NA L-lactate permease Carbohydrates, organic alcohols, and acids -1.87 0.00 -1.71 -2.27 8.89 0.00 SA2437 bcr bicyclomycin resistance protein Other -1.86 0.00 0.00 -3.75 -1.84 0.00 SA1994 NA ABC transporter, ATP-binding Unknown substrate -1.85 0.00 0.00 -3.99 -3.87 0.00 protein SA2138 NA cation efflux family protein Cations and iron carrying compounds -1.83 0.00 0.00 0.00 0.00 0.00 SA0195 NA maltose ABC transporter, permease Carbohydrates, organic alcohols, and acids -1.82 -2.44 0.00 0.00 -6.82 0.00 protein SA2441 NA amino acid permease Amino acids, peptides and amines -1.80 0.00 0.00 -4.61 -6.14 1.63 SA1384 opuD1 osmoprotectant transporter, BCCT Amino acids, peptides and amines -1.80 0.00 0.00 0.00 -1.89 0.00 family SA0010 NA AzlC family protein Amino acids, peptides and amines -1.79 0.00 0.00 0.00 0.00 0.00 SA2460 NA drug transporter, putative Other -1.74 0.00 0.00 -2.97 0.00 0.00 SA2257 NA drug transporter, putative Other -1.73 0.00 0.00 -3.10 0.00 0.00 SA0098 sirB iron compound ABC transporter, Cations and iron carrying compounds -1.71 0.00 0.00 -2.35 0.00 0.00 permease protein SirB 15
SA2314 NA sodium/bile acid symporter family Other -1.70 0.00 0.00 0.00 0.00 0.00 protein SA1392 NA sodium:alanine symporter family Amino acids, peptides and amines -1.67 -1.60 -1.86 -2.57 -2.15 1.72 protein SA0788 NA proton-dependent oligopeptide Amino acids, peptides and amines -1.66 0.00 -1.88 0.00 -2.13 0.00 transporter family protein SA2167 NA iron compound ABC transporter, Cations and iron carrying compounds -1.63 0.00 -1.98 2.29 0.00 0.00 iron compound-binding protein SA1211 uraA uracil permease Nucleosides, purines and pyrimidines -1.58 0.00 0.00 0.00 0.00 2.08 SA1963 NA proline permease Amino acids, peptides and amines -1.57 0.00 0.00 2.18 0.00 0.00 SA2347 NA drug resistance transporter, Other -1.57 0.00 -1.66 -1.72 0.00 1.56 EmrB/QacA subfamily SA0097 sirC iron compound ABC transporter, Cations and iron carrying compounds -1.56 0.00 0.00 -2.66 0.00 0.00 permease protein SirC SA0516 NA PTS system, IIBC components Carbohydrates, organic alcohols, and acids -1.55 0.00 0.00 -3.25 0.00 -1.57 SA2181 lacE PTS system, lactose-specific IIBC Carbohydrates, organic alcohols, and acids -1.52 0.00 0.00 1.64 0.00 0.00 components SA2663 NA PTS system, fructose-specific Carbohydrates, organic alcohols, and acids -1.51 0.00 0.00 0.00 -3.85 0.00 IIABC components SA2348 NA drug transporter, putative Other -1.51 0.00 -1.25 0.00 0.00 0.00 SA2316 NA PTS system, IIBC components Carbohydrates, organic alcohols, and acids -2.43 -2.64 0.00 -8.25 -3.83 0.00 SA0175 NA PTS system, IIABC components Carbohydrates, organic alcohols, and acids -2.27 0.00 -1.93 -3.21 0.00 0.00 SA2552 NA PTS system, IIABC components Carbohydrates, organic alcohols, and acids -2.14 0.00 0.00 -2.13 0.00 0.00 SA2376 NA PTS system, sucrose-specific IIBC Carbohydrates, organic alcohols, and acids -1.88 -1.81 0.00 -10.57 -2.24 -1.61 components, putative SA0516 NA PTS system, IIBC components Carbohydrates, organic alcohols, and acids -1.55 0.00 0.00 -3.25 0.00 -1.57 SA2181 lacE PTS system, lactose-specific IIBC Carbohydrates, organic alcohols, and acids -1.52 0.00 0.00 1.64 0.00 0.00 components SA2663 NA PTS system, fructose-specific Carbohydrates, organic alcohols, and acids -1.51 0.00 0.00 0.00 -3.85 0.00 IIABC components SA0193 NA maltose ABC transporter, maltose- Carbohydrates, organic alcohols, and acids -2.68 -3.66 0.00 -1.81 -5.93 0.00 binding protein, putative SA1045 NA iron compound ABC transporter, Cations and iron carrying compounds -2.11 0.00 0.00 0.00 0.00 0.00 iron compound-binding protein SA2167 NA iron compound ABC transporter, Cations and iron carrying compounds -1.63 0.00 -1.98 2.29 0.00 0.00 iron compound-binding protein SA2437 bcr bicyclomycin resistance protein Other -1.86 0.00 0.00 -3.75 -1.84 0.00 16
SA2460 NA drug transporter, putative Other -1.74 0.00 0.00 -2.97 0.00 0.00 SA2257 NA drug transporter, putative Other -1.73 0.00 0.00 -3.10 0.00 0.00 SA2347 NA drug resistance transporter, Other -1.57 0.00 -1.66 -1.72 0.00 1.56 EmrB/QacA subfamily
Hypothetical proteins
SA1086 NA conserved hypothetical protein Conserved -2.87 0.00 0.00 0.00 0.00 2.22 SA1136 NA conserved hypothetical protein Conserved -2.80 0.00 0.00 -2.91 -2.56 1.93 SA1044 NA conserved hypothetical protein Conserved -2.77 0.00 -1.97 -2.43 0.00 0.00 SA1307 NA conserved hypothetical protein Conserved -2.62 0.00 -1.63 -2.03 0.00 0.00 TIGR00282 SA1017 NA conserved hypothetical protein Conserved -2.59 -1.61 -2.08 -3.46 -2.03 0.00 SA2334 NA conserved hypothetical protein Conserved -2.52 0.00 -2.14 -3.16 -1.61 0.00 SA0755 NA conserved hypothetical protein Conserved -2.48 0.00 -2.21 -3.61 0.00 0.00 SA0271 NA conserved hypothetical protein Conserved -2.38 0.00 0.00 -1.57 0.00 0.00 SA1664 NA conserved hypothetical protein Conserved -2.38 0.00 1.94 -2.03 2.47 0.00 TIGR00370 SA0643 NA conserved hypothetical protein Conserved -2.34 0.00 0.00 0.00 0.00 0.00 SA0711 NA conserved hypothetical protein Conserved -2.25 0.00 1.56 -7.91 -1.59 2.11 SA1935 NA conserved hypothetical protein Conserved -2.10 0.00 0.00 0.00 0.00 0.00 SA2373 NA conserved hypothetical protein Conserved -2.09 -1.83 0.00 -4.08 0.00 0.00 SA0067 NA conserved hypothetical protein Conserved -2.03 0.00 0.00 -2.77 0.00 0.00 SA1991 NA conserved hypothetical protein Conserved -2.00 0.00 0.00 -3.36 0.00 0.00 SA1701 NA conserved hypothetical protein Conserved -2.00 0.00 0.00 -5.10 0.00 0.00 SA1126 NA conserved hypothetical protein Conserved -1.99 0.00 0.00 -2.96 0.00 0.00 SA1993 NA conserved hypothetical protein Conserved -1.97 0.00 0.00 -5.25 0.00 0.00 SA1481 NA conserved hypothetical protein Conserved -1.97 0.00 0.00 -1.93 0.00 0.00 SA1805 NA ORFID:SA1805~repressor Conserved -1.96 0.00 0.00 0.00 0.00 0.00 homolog [Bacteriophage phiN315] SA0281 NA conserved hypothetical protein Conserved -1.95 0.00 0.00 0.00 0.00 0.00 SA1297 NA conserved hypothetical protein Conserved -1.93 0.00 0.00 -3.59 0.00 0.00 SA1947 NA conserved hypothetical protein Conserved -1.91 0.00 0.00 -1.79 0.00 0.00 SA0599 NA conserved hypothetical protein Conserved -1.88 0.00 0.00 -10.70 -1.83 0.00 SA0565 NA conserved hypothetical protein Conserved -1.88 -1.75 -1.58 -3.62 -1.89 0.00 SA0059 NA conserved hypothetical protein Conserved -1.87 0.00 0.00 -4.22 0.00 0.00 SA1426 NA conserved hypothetical protein Conserved -1.87 -1.61 0.00 0.00 0.00 0.00 17
SA1112 NA conserved hypothetical protein Conserved -1.82 0.00 0.00 -1.96 0.00 0.00 SA1359 NA conserved hypothetical protein Conserved -1.73 0.00 0.00 -2.66 0.00 0.00 SA2587 NA conserved hypothetical protein Conserved -1.72 0.00 0.00 0.00 0.00 0.00 SA0011 NA conserved hypothetical protein Conserved -1.71 0.00 0.00 0.00 0.00 0.00 SA0628 NA conserved hypothetical protein Conserved -1.67 0.00 0.00 0.00 0.00 0.00 SA1373 NA conserved hypothetical protein Conserved -1.66 -1.76 1.79 -6.20 -2.89 2.05 SA1388 NA conserved hypothetical protein Conserved -1.66 0.00 0.00 -1.91 -2.67 0.00 TIGR00023 SA1840 NA conserved hypothetical protein Conserved -1.64 0.00 0.00 -2.16 0.00 0.00 SA2134 NA conserved hypothetical protein Conserved -1.64 0.00 0.00 -2.46 0.00 0.00 SA1989 NA conserved hypothetical protein Conserved -1.64 0.00 0.00 -1.80 0.00 0.00 SA2661 NA conserved hypothetical protein Conserved -1.62 0.00 0.00 0.00 0.00 0.00 SA1990 NA conserved hypothetical protein Conserved -1.60 0.00 0.00 -1.98 0.00 0.00 SA1995 NA conserved hypothetical protein Conserved -1.59 0.00 0.00 -3.92 0.00 0.00 SA0974 NA conserved hypothetical protein Conserved -1.59 0.00 0.00 0.00 0.00 0.00 SA0198 NA conserved hypothetical protein Conserved -1.59 -2.63 0.00 0.00 -3.84 0.00 SA2601 NA conserved hypothetical protein Conserved -1.57 0.00 0.00 0.00 0.00 0.00 SA0662 NA conserved hypothetical protein Conserved -1.57 0.00 -1.50 -1.67 0.00 0.00 SA0489 NA conserved hypothetical protein Conserved -1.57 0.00 0.00 0.00 0.00 0.00 SA1968 NA conserved hypothetical protein Conserved -1.56 0.00 0.00 -1.71 0.00 0.00 SA2118 NA conserved hypothetical protein Conserved -1.54 0.00 0.00 0.00 0.00 0.00 SA0529 NA conserved hypothetical protein Conserved -1.54 0.00 0.00 -1.55 0.00 0.00 SA0527 NA conserved hypothetical protein Conserved -1.53 0.00 0.00 -3.38 0.00 0.00 SA2672 NA conserved hypothetical protein Conserved -1.53 0.00 0.00 -1.98 0.00 0.00 SA0528 NA conserved hypothetical protein Conserved -1.53 0.00 0.00 0.00 0.00 0.00 SA2300 NA conserved hypothetical protein Conserved -1.52 0.00 -1.93 1.55 0.00 0.00 SA1763 NA conserved hypothetical protein Conserved -1.51 0.00 0.00 0.00 0.00 0.00 SA2557 NA conserved domain protein Domain -4.03 0.00 0.00 -5.53 0.00 1.96 SA2037 NA conserved domain protein Domain -2.56 0.00 0.00 -4.04 0.00 0.00 SA1847 NA conserved domain protein, putative Domain -1.98 0.00 0.00 0.00 0.00 0.00
Unknown function
SA2536 NA hydrolase, haloacid dehalogenase- Enzymes of unknown specificity -3.03 -2.94 0.00 -2.60 -2.31 0.00 like family SA2297 NA monooxygenase family protein Enzymes of unknown specificity -2.46 0.00 -1.83 -2.02 0.00 1.75 SA0196 NA oxidoreductase, Gfo/Idh/MocA Enzymes of unknown specificity -2.19 -2.69 0.00 0.00 -9.04 0.00 18
family SA2245 NA acetyltransferase, GNAT family Enzymes of unknown specificity -1.76 0.00 0.00 0.00 0.00 0.00 SA0606 NA hydrolase, haloacid dehalogenase- Enzymes of unknown specificity -1.74 0.00 0.00 -2.17 0.00 0.00 like family SA2541 NA acetyltransferase, GNAT family Enzymes of unknown specificity -1.70 0.00 0.00 0.00 0.00 0.00 SA0197 NA oxidoreductase, Gfo/Idh/MocA Enzymes of unknown specificity -1.68 -2.11 0.00 1.78 -2.52 0.00 family SA0605 NA cytidine/deoxycytidylate deaminase Enzymes of unknown specificity -1.68 0.00 0.00 0.00 0.00 0.00 family protein, authentic frameshift SA2293 NA NAD/NADP octopine/nopaline Enzymes of unknown specificity -1.63 0.00 -2.25 -6.95 0.00 0.00 dehydrogenase family protein SA0507 NA LysM domain protein General -3.05 -1.59 -2.59 -4.46 0.00 4.61 SA2109 NA modification methylase, HemK General -2.33 -2.07 0.00 -5.21 -1.54 0.00 family SA1548 NA AtsA/ElaC family protein General -2.27 0.00 0.00 -3.27 0.00 0.00 SA0723 NA LysM domain protein General -2.17 0.00 -2.41 -4.09 -3.00 2.34 SA2328 NA abortive infection protein family General -2.14 0.00 -2.07 -3.44 -3.95 2.43 SA1118 typA GTP-binding protein TypA General -2.14 -1.77 0.00 -7.18 0.00 2.52 SA1186 NA antibacterial protein (phenol soluble General -2.07 0.00 0.00 2.92 0.00 0.00 modulin) SA0812 NA degV family protein General -2.06 -1.82 0.00 -1.68 -2.04 0.00 SA1187 NA antibacterial protein (phenol soluble General -2.03 0.00 0.00 5.18 0.00 0.00 modulin) SA0276 yukA diarrheal toxin General -1.99 0.00 0.00 -1.62 0.00 0.00 SA0772 NA exsB protein General -1.99 0.00 0.00 -2.13 0.00 0.00 SA0007 NA YjeF-related protein General -1.92 0.00 0.00 0.00 0.00 0.00 SA0553 tilS MesJ/Ycf62 family protein General -1.84 0.00 0.00 -4.15 0.00 0.00 SA0021 yycH yycH protein General -1.78 1.93 0.00 -1.65 0.00 0.00 SA2275 NA BioY family protein General -1.76 0.00 0.00 0.00 0.00 0.00 SA1807 NA rhodanese-like domain protein General -1.69 0.00 0.00 0.00 0.00 0.00 SA0777 NA urea amidolyase-related protein General -1.67 -1.63 0.00 0.00 0.00 0.00 SA2333 NA YnfA family protein General -1.67 0.00 -1.54 1.91 0.00 0.00 SA0790 NA integral membrane domain protein General -1.63 0.00 0.00 -2.17 0.00 0.00 SA1720 NA GTP-binding protein, putative General -1.62 0.00 0.00 0.00 0.00 0.00 SA2080 NA HD domain protein General -1.58 0.00 0.00 -2.22 0.00 0.00 19 a Fold value refers to expression increases or decreases for up- or down-regulated genes respectively. b 0.00 indicates data not available or no change. c Dap – Daptomycin; Van – Vancomycin; Oxa – Oxacillin; CCCP - carbonyl cyanide m-chlorophenylhydrazone; Nis – Nisin. d KVR – S. aureus vraSR- mutant KVR derived from S. aureus N315 (31).