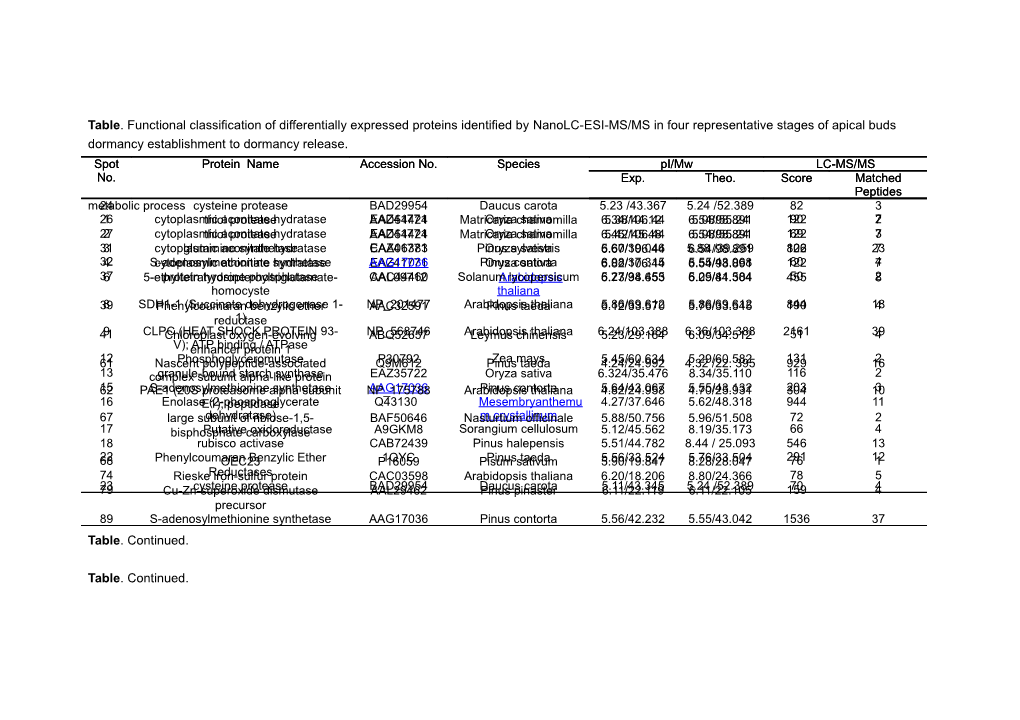
Table. Functional classification of differentially expressed proteins identified by NanoLC-ESI-MS/MS in four representative stages of apical buds dormancy establishment to dormancy release. Spot Protein Name Accession No. Species pI/Mw LC-MS/MS No. Exp. Theo. Score Matched Peptides metabolic24 process cysteine protease BAD29954 Daucus carota 5.23 /43.367 5.24 /52.389 82 3 261 cytoplasmicthiol aconitate protease hydratase AAD54424EAZ41771 MatricariaOryza chamomilla sativa 6.34/106.445.38/44.12 6.54/5.08/55.298.8914 12290 72 272 cytoplasmicthiol aconitate protease hydratase AAD54424EAZ41771 MatricariaOryza chamomilla sativa 6.45/106.445.42/45.48 6.54/5.08/55.298.8914 12269 73 313 cytoplasmicglutamine aconitate synthetase hydratase CAA06383EAZ41771 PinusOryza sylvestris sativa 6.67/106.445.60/39.046 5.886.54/ /39.25998.891 122806 237 324 S-adenosylmethioninecytoplasmic aconitate synthetasehydratase AAG17036EAZ41771 PinusOryza contorta sativa 6.82/106.446.08/37.315 6.54/5.55/43.06898.891 12260 74 376 5-ethyltetrahydropteroyltriglutamate-protein tyrosine phosphatase CAC44460AAL09712 SolanumArabidopsis lycopersicum 6.73/94.6535.27/38.455 6.09/84.3045.25/41.584 45550 82 homocyste thaliana 398 SDH1-1Phenylcoumaran (Succinate dehydrogenase benzylic ether 1- NP_201477AAC32591 ArabidopsisPinus taeda thaliana 6.5.86/69.61212/33.570 5.76/33.55.86/69.61245 190844 184 reductase1) 419 CLPCChloroplast (HEAT SHOCK oxygen-evolving PROTEIN 93- NP_568746ABQ52657 ArabidopsisLeymus chinensis thaliana 6.24/103.3885.23/29.164 6.36/103.3886.09/34.512 216151 394 V); enhancerATP binding protein / ATPase 1 6112 NascentPhosphoglyceromutase polypeptide-associated Q9M612P30792 PinusZea mays taeda 4.24/24.9925.45/60.634 4.325.29/60. /22.582 395 929131 162 13 complexgranule-bound subunit starchalpha-like synthase protein EAZ35722 Oryza sativa 6.324/35.476 8.34/35.110 116 2 6215 PAE1S-adenosylmethionine (20S proteasome alpha synthetase subunit NP_175788AAG17036 ArabidopsisPinus contorta thaliana 4.82/24.9585.64/43.067 4.70/25.9315.55/43.132 304203 103 16 EnolaseE1); (2-phosphoglycerate peptidase Q43130 Mesembryanthemu 4.27/37.646 5.62/48.318 944 11 67 large subunitdehydratase) of riblose-1,5- BAF50646 Nasturtiumm crystallinum officinale 5.88/50.756 5.96/51.508 72 2 17 bisphosphatePutative carboxylase oxidoreductase A9GKM8 Sorangium cellulosum 5.12/45.562 8.19/35.173 66 4 18 rubisco activase CAB72439 Pinus halepensis 5.51/44.782 8.44 / 25.093 546 13 6822 PhenylcoumaranOEC23 Benzylic Ether P160591QYC PisumPinus sativum taeda 5.90/19.8475.56/33.524 8.28/28.0475.76/33.504 29176 121 74 RieskeReductases iron-sulfur protein CAC03598 Arabidopsis thaliana 6.20/18.206 8.80/24.366 78 5 7923 Cu-Zn-superoxidecysteine protease dismutase BAD29954AAL29462 DaucusPinus pinaster carota 5.11/43.3456.11/22.119 5.246.11/22. /52.389105 15970 4 precursor 89 S-adenosylmethionine synthetase AAG17036 Pinus contorta 5.56/42.232 5.55/43.042 1536 37 Table. Continued.
Table. Continued. Table. Continued. Spot Protein Name Accession No. Species pI/Mw LC-MS/MS No. Exp. Theo. Score Matched Peptides 7091 S-adenosylmethionineSuperoxide dismutase synthetase [Cu-Zn], 1 P24707P49611 PinusBrassica sylvestris juncea 6.07/19.5265.91/33.234 5.16/14.4265.52/43.199 306173 45 92 beta-hydroxyacyl-ACPchloroplast dehydratase ABA25920 Picea mariana 5.93/22.781 9.03/26.821 154 3 7194 17.5PSI-E kDa subunitclass I ofheat photosystem shock protein I BAA07667Q69BI7 NicotianaCarica papayasylvestris 6.05/16.61/18.2247.481 5.32/17.49.74/15.59971 15859 24 Cellular72 processeslow molecular weight heat-shock CAA63570 Pseudotsuga menziesii 6.10/18.201 5.82/18.180 45 2 5 719 23 Cell divisionprotein cycle protein 48 AAA80587 soybean 5.23/88.765 5.18/89.714 29 54 1 73 lowProtein molecular disulfide weight isomerase heat-shock CAA63570CAA72092 PseudotsugaNicotiana tabacum menziesii 6.12/5.69/4 18.2300.645 5.82/5.99/3 18.1809.683 258 7 34 two-componentprotein hybrid sensor and ZP_01549722 Stappia aggregata IAM 6.58/40.876 4.77/41.506 47 3 76 intracellular pathogenesis-relatedregulator AAC33531 Pinus monticola 6.55/18.000 5.56/17.99 239 27 38 Abscisic acid-andprotein PinmIII stress-induced AAV92173 Pseudotsuga menziesii 5.33/37.378 4.36/12.369 56 1 77 intracellular pathogenesis-relatedprotein AAC33531 Pinus monticola 6.56/17.625 5.56/17.99 243 4 40 Abscisicprotein acid and PinmIII water-stress B7TGP2 Pinus sylvestris 5.43/30.212 9.55/16.292 43 2 78 inducedPR10 protein protein AAL50006 Pinus monticola 6.13/17.791 5.30/17.780 95 1 8745 ascorbateCatalase peroxidase ZP_01227619AAR32786 AurantimonasPinus pinaster sp. 5.25/19.4555.34/27.510 5.54/78.4155.40/27.520 29642 27 Protein46 synthesis ascorbate peroxidase ABK23188 Pinus pinaster 5.13/27.450 5.40/27.250 207 4 1947 AbscisicEukaryotic acid-and initiation stress-induced factor 4A-15 AAW59260Q40468 NicotianaPinus taedatabacum 5.72/46.1785.15/25.56 8.444.83/10.999 / 46.687 38846 82 20 eukaryotic initiationprotein factor 4A BAA02152 Oryza sativa Japonica 5.89/45.157 5.29 /46.902 649 14 48 Abscisic acid-and stress-induced AAW59260 PinusGroup taeda 5.18/26.549 4.83/10.999 54 4 21 chloroplast translationalprotein elongation P46280 Pelargonium 5.45/52.809 6.33/52.467 251 9 49 Abscisic acid-andfactor stress-inducedTu AAW59260 Pinusgraveolens taeda 5.19/25.921 4.83/10.999 50 4 54 40S ribosomalprotein protein S3 (RPS3A) NP_180719 Arabidopsis thaliana 6.12/ 26.502 9.57/ 27.502 186 3 5055 40S ribosomalascorbate protein peroxidase S3 (RPS3A) NP_180719AAR32786 ArabidopsisPinus pinaster thaliana 6.47/5.20/27.221 26.532 9.57/5.40/27.520 27.502 464145 113 83 40S ribosomal protein S12 ABK22790 Picea sitchensis 5.4/17.964 5.32/14.954 424 14 51 Abscisic acid-and stress-induced AAW59260 Pinus taeda 5.21/26.999 4.83/10.999 50 3 protein 52 Abscisic acid-and stress-induced AAW59260 Pinus taeda 5.22/27.08 4.83/10.999 64 3 protein
Table. Continued. Table. Continued. Spot Protein Name Accession No. Species pI/Mw LC-MS/MS No. Exp. Theo. Score Matched Peptides 8435 CarbonicPutative uncharacterized anhydrase, chloroplast protein ABK25140P27141 NicotianaPicea sitchensis tabacum 5.14/41.9815.91/19.5 5.456.4/34.489 /43.795 1266641 4020 43 precursor (Carbonateunknown dehydratase) ABK21186 Picea sitchensis 5.01/24.039 4.99/23.019 381 11 Structural53 componentUnknown protein ACN39903 Picea sitchensis 6.25/27.504 6.43/48.678 32 1 5625 unknownactin ABK22547AAF03692 PiceaPicea sitchensis rubens 5.59/5.35/43.543 26.390 5.58/5.30 /41.56426.690 244475 124 5730 type IIIa membraneUnknown protein cp-wap13 AAB61672ABK23407 VignaPicea unguiculata sitchensis 5.12/5.56/38.893 27.219 8.39/6.24 /39.39627.198 824997 2233 5836 Unknownactin ABK23835AAF03692 PiceaPicea sitchensis rubens 4.17/23.0785.21/39.647 4.325.30 /22.485/41.564 586553 1812 5942 Caffeoyl-CoAunknown O-methyltransferase ABK21546Q9ZTT5 PiceaPinus sitchensis taeda 4.23/27.1775.35/29.127 4.255.43/29.127 /22.904 463760 1223 60 (Trans-caffeoyl-CoAunknown ABK21546 Picea sitchensis 4.18/22.054 4.25 /22.904 214 3 63 3-O-methyltransferase)unknown (CCoAMT) ABK23835 Picea sitchensis 4.33/21.425 4.32/22.485 237 3 64 (CCoAOMT)unknown ABK24055 Picea sitchensis 4.71/22.352 5.04/32.352 186 10 6544 Caffeoyl-CoAunknown O-methyltransferase ABK24573Q9ZTT5 PiceaPinus sitchensis taeda 4.79/22.2505.12/29.932 5.434.88/31.249 /29.127 18699 3 66 (Trans-caffeoyl-CoAunknown ABK25464 Picea sitchensis 5.38/22.445 6.10/25.565 364 6 69 3-O-methyltransferase)unknown (CCoAMT) ABK25464 Picea sitchensis 6.01/19.934 6.10/25.565 174 3 75 (CCoAOMT)unknown ABK26804 Picea sitchensis 6.22/18.234 6.86/18.250 177 4 Unknown81 or Putativehypothetical uncharacterized protein protein A9NU54 Picea sitchensis 5.82/16.257 5.06/16.777 42 2 827 unnamedhypothetical protein protein product CAO16769CAO68394 Vitis vinifera 5.23/17.2415.42/83.427 6.195.91/76.282 / 42.694 565701 139 8510 Putativehypothetical uncharacterized protein protein CAN60522ABK22623 PiceaVitis sitchensisvinifera 6.583/73.7685.95/19.231 9.71/12.4416.63/73.732 21395 42 8611 Putativehypothetical uncharacterized protein protein CAN74538A0NLS3 LabrenziaVitis vinifera aggregata 5.93/19.1746.12/64.867 4.93/22.8846.07/64.936 4244 2 1490 hypotheticalunknown protein CAN77028ABK23829 PiceaVitis sitchensisvinifera 6.527/5.45/31.23553.167 7.57/53.5.31/31.628150 34198 68 2893 Putative uncharacterizedunknown protein ABK26037A0NLS4 PiceaPicea sitchensis rubens 5.64/42.0456.25/19.897 5.55/44.0428.48/20.902 25047 26 33 hypothetical protein EAZ41123 Oryza sativa 6.49/38.7 7.29/55.734 249 7
Table . Continued. Spot Protein Name Accession No. Species pI/Mw LC-MS/MS No. Exp. Theo. Score Matched Peptides 95 unknown ABK21236 Picea sitchensis 4.19/37.543 5.75/40.965 216 6 Others 80 glycine-rich RNA-binding protein Q9XEL4 Picea glauca 6.10/15.450 7.85/15.441 186 3 88 Cystatin P25973 Glycine max 6.13/18.273 6.78/5.274 54 1 96 Peptidase T1A, proteasome beta- ABE90866 Medicago truncatula 5.79/ 27.860 5.50/ 28.773 350 12 subunit