Supplementary Figures
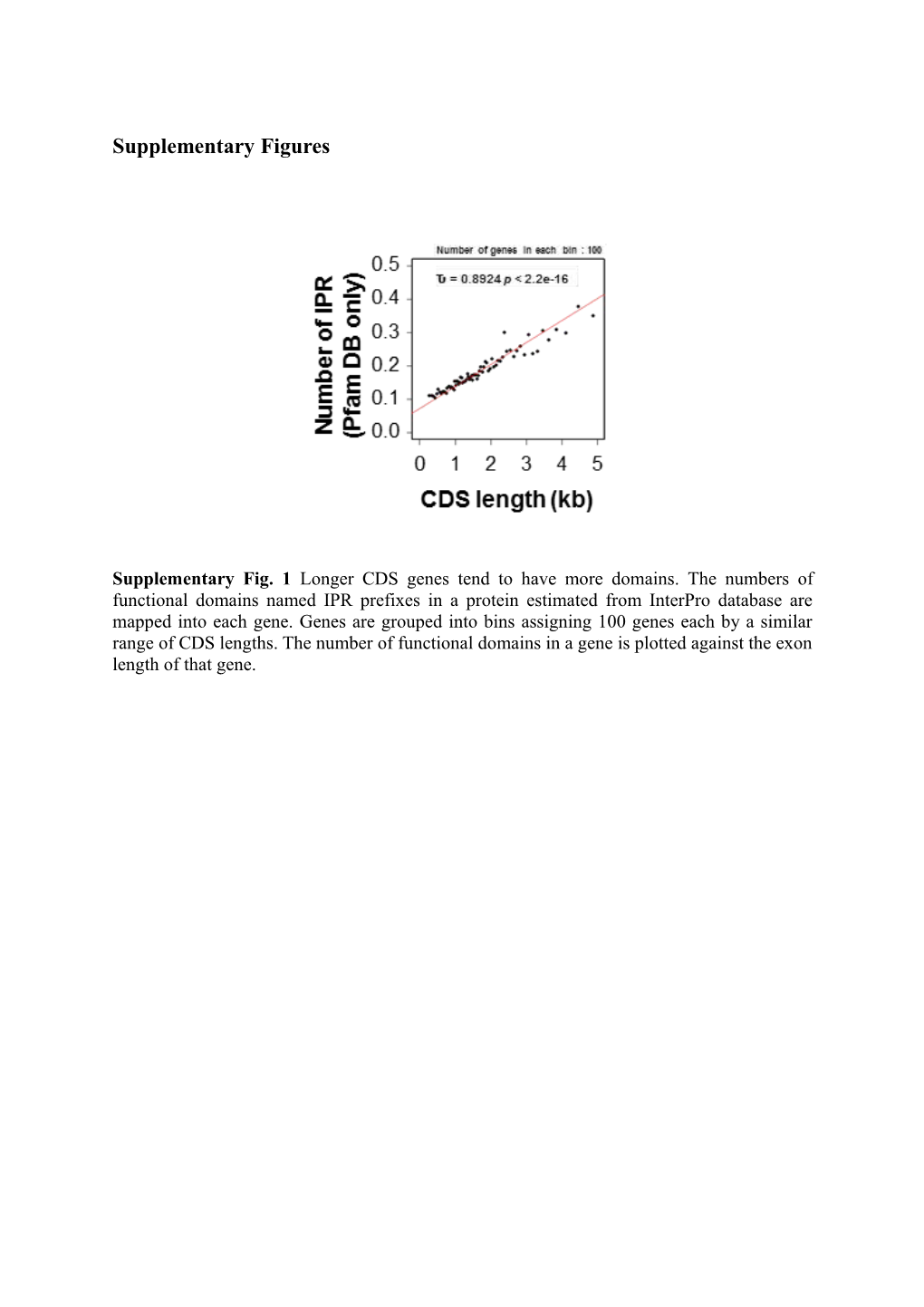
Supplementary Fig. 1 Longer CDS genes tend to have more domains. The numbers of functional domains named IPR prefixes in a protein estimated from InterPro database are mapped into each gene. Genes are grouped into bins assigning 100 genes each by a similar range of CDS lengths. The number of functional domains in a gene is plotted against the exon length of that gene. Supplementary Fig. 2 Lethal genes tend to have longer CDS length and evolve slower than non-lethal genes in yeast. a Please refer to the legends of Figure 1b. Scatter plot of the CDS length groups versus proportion of lethal genes for each length group in yeast. b Comparison of dN values between the ‘essential genes’ group and the ‘nonessential genes’ group after genes were divided by their lethality information into two groups in yeast. Supplementary Fig. 3 Comparison of dN/dS in different sizes of each subcomponent length groups. The same analysis as done for Figure 2a except dN/dS instead of dN values were used. Please refer to the legends of Figure 2a. Supplementary Fig. 4 Negative relationship between dN/dS and proportion of conserved sites in CDS. Conserved sites in the CDS regions were estimated by the threshold of a PhastCons score ≥ 0.5, as the same way as done for Figure 1d. dN/dS ratios were estimated using human/mouse orthologs (see Materials and methods) for each gene with a certain length of CDS. The two values were plotted against each other, and Kendall’s correlation test was performed. Supplementary Tables
Supplementary Table 1 Kendall’s correlation analysis among the lengths of different subcomponent in a gene
ԏ Human ԏ Mouse 5` UTR 3` UTR CDS 5` UTR 3` UTR CDS
3` UTR 0.1153 - - 0.1823 - -
CDS 0.0306 0.0876 - 0.0959 0.1095 -
Intron 0.1205 0.2162 0.2919 0.1725 0.2135 0.2825