PRIMER DESIGN – OVERVIEW
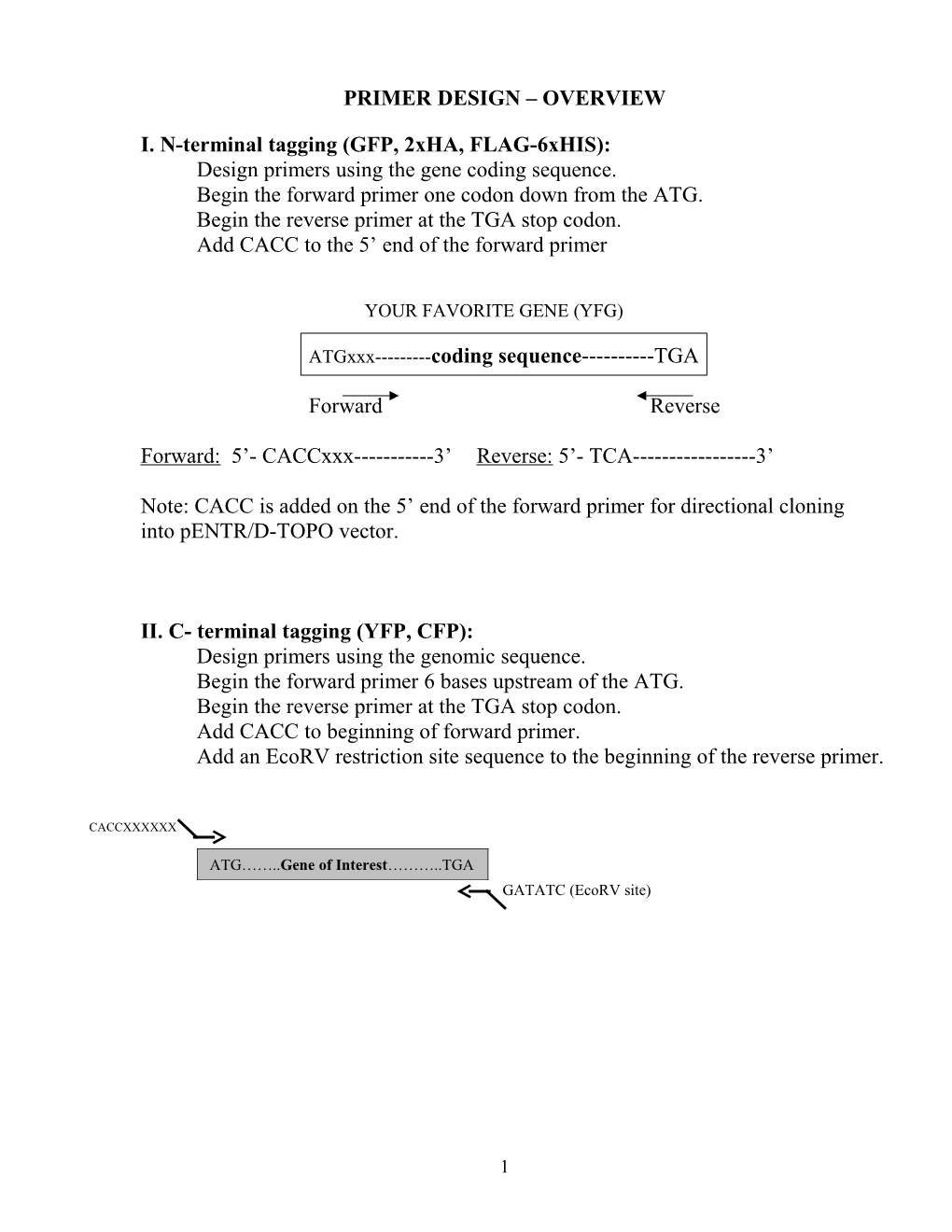
I. N-terminal tagging (GFP, 2xHA, FLAG-6xHIS): Design primers using the gene coding sequence. Begin the forward primer one codon down from the ATG. Begin the reverse primer at the TGA stop codon. Add CACC to the 5’ end of the forward primer
YOUR FAVORITE GENE (YFG)
ATGxxx------coding sequence------TGA
Forward Reverse
Forward: 5’- CACCxxx------3’ Reverse: 5’- TCA------3’
Note: CACC is added on the 5’ end of the forward primer for directional cloning into pENTR/D-TOPO vector.
II. C- terminal tagging (YFP, CFP): Design primers using the genomic sequence. Begin the forward primer 6 bases upstream of the ATG. Begin the reverse primer at the TGA stop codon. Add CACC to beginning of forward primer. Add an EcoRV restriction site sequence to the beginning of the reverse primer.
CACCXXXXXX
ATG……..Gene of Interest………..TGA GATATC (EcoRV site)
1 III. Knock-out construction Design primers using the genomic sequence. Design a primer set to amplify the 5’ flanking region Design a primer set to amplify the 3’ flanking region
YOUR FAVORITE GENE (YFG) 5’ flanking 3’ flanking ATGxxx------coding sequence------TGA
5’ Fl forward 5’ Fl reverse 3’ Fl forward 3’ Fl Reverse
IV. Gene Expression Analysis Design primers using the genomic sequence. The forward and reverse primers must be on either side of at least one intron. The introns(s) flanked should add up to at least 100 bp The exonic sequence between the two primers should be 400-600 bps.
Example:
ATG- Exon 1 Exon 2 Exon 3 Exon 4
Forward Reverse
2