Supporting Information
Combining Molecular Dynamics Simulation And Ligand-Receptor Contacts
Analysis As A New Approach For Pharmcophore Modeling: Beta-Secretase 1
And Check Point Kinase 1 As Case Studies
Ma'mon M. Hatmala, Shadi Jaberb, Mutasem O. Tahac*
aDepartment of Medical Laboratory Sciences, Faculty of Allied Health Sciences, The
Hashemite University, Zarqa, Jordan
bDepartment of Medical Laboratory Sciences, Faculty of Allied Health Sciences, Zar-
qa Private University, Zarqa, Jordan
cDrug Discovery Unit, Department of Pharmaceutical Sciences, Faculty of Pharmacy,
University of Jordan, Amman, Jordan
*Corresponding Author,
Telephone: 00962 6 5355000 ext. 23305.
Fax: 00962 6 5339649.
Email: [email protected]
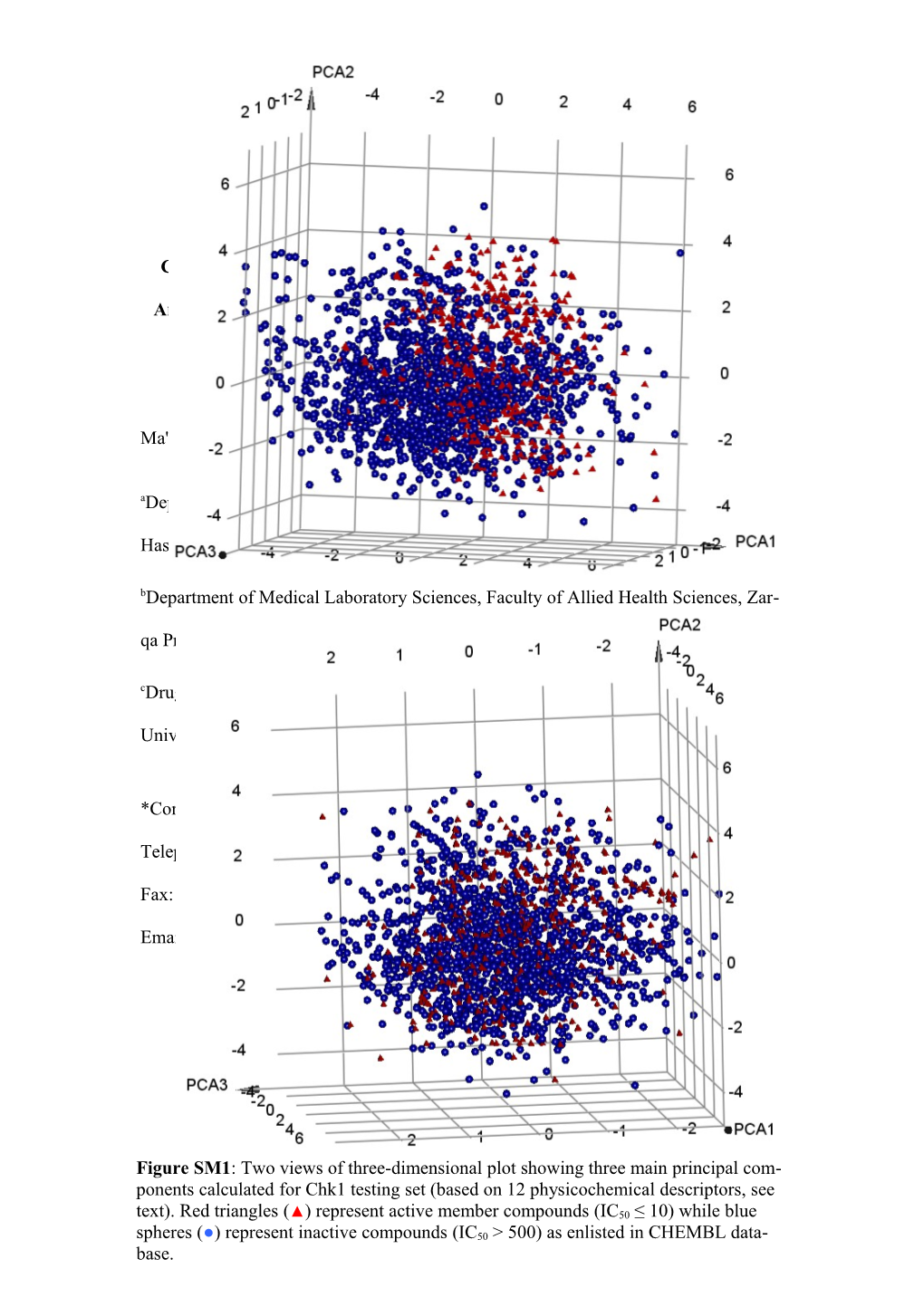
Figure SM1: Two views of three-dimensional plot showing three main principal com- ponents calculated for Chk1 testing set (based on 12 physicochemical descriptors, see text). Red triangles (▲) represent active member compounds (IC50 ≤ 10) while blue spheres (●) represent inactive compounds (IC50 > 500) as enlisted in CHEMBL data- base.
Figure SM2: Two views of three-dimensional plot showing three main principal com- ponents calculated for BACE1 testing set (based on 12 physicochemical descriptors, see text). Red triangles (▲) represent active member compounds (IC50 ≤ 10) while blue spheres (●) represent inactive compounds (IC50 > 500) as enlisted in CHEMBL data- base. Section SM-1: ROC analysis
ROC curve is plotted by considering the highest score of an active molecule as the
first threshold then counting the number of inactive compounds within this cut-off
value, and both the corresponding sensitivity (Se) and specificity (Sp) are calculated
using equation 1 and equation 2, respectively. This process is repeated using the ac-
tive molecule possessing the second highest score and so on, until the scores of all ac-
tive compounds are considered as selection cut-off values [41].
Number of Selected Actives TP Se ………………….. (1) Total Number of Actives TP FN
….…….……….. (2) Number of Discarded Inactives TN Sp Total Number of Inactives TN FP where, TP (true positive) is the number of active compounds that are captured by the
pharmacophore under concern, FN (false negative) is the number of active com-
pounds discarded from the hits list, TN (true negative) is the number of discarded de-
coys , while FP (false positive) is the number of captured decoys .
The ROC curve for ideal distributions, where active compounds are completely sepa-
rate and distinct from the inactives (i.e., no overlap between actives and decoys), the
curve arise vertically to the upper-left corner (Se = Sp = 1) and then joins the up-
per-right corner horizontally. Hence, the more a ROC curve bends towards the upper
left corner of the diagram, the more distinct the signal appears [41].
In practice, the ROC curve for a set of actives and decoys with randomly distributed
scores tends towards the Se = 1-Sp line asymptotically with increasing number of ac-
tives and decoys.33 The success of particular virtual screening workflow depending
on ROC analysis evaluation can be provided as follow: 1) Area under the ROC curve (AUC): which range from 0.5 to 1, where 1 indicates an optimal value and 0.5 random distribution. Whereas an AUC value lower than 0.5 represents the unfavourable case of a virtual screening method that has a higher prob- ability to assign the best scores to decoys than to actives [41].
2) Overall specificity (SPC): that describes the percentage of discarded inactive by the particular virtual screening workflow. Inactive test compounds are assigned a binary score value of zero (compound not captured) or one (compound captured) [41-43].
3) Overall true positive rate (TPR or overall sensitivity): describes the fraction per- centage of captured actives from the total number of actives. Active test compounds are assigned a binary score value of zero (compound not captured) or one (compound captured) [41-43].